(Now clickable!)
|
1. EMBL Meyerhofstrasse 1 D-69012, Heidelberg Germany Home Page |
2. DeCODE Genetics Bioinformatics Sturlugoetu 8 101 Reykjavik Iceland |
3. GlaxoSmithKline Bioinformatics NFSP-N, 3rd Avenue Harlow, Essex CM19 5AW U.K. |
Much of what these pages contain is a perspective from one person. Probably I give myself too much credit regarding my knowledge of the principles of protein structure and function, and publications and such like have gone to my head. So please read these pages with these caveats in mind. I don't know everything, and this is very much a perspective based on my own experience and knowledge. Having said that, I have looked at a lot of protein sequences and structures in my time, so I anticipate and hope the pages will be of some use to anyone who doesn't know all that much about structure and (for example) is wondering what the consequence of a particular mutation might be.
Details like substitution or active site preferences are derived systematically, so can be trusted within the limits of how they are compiled. There are many thousands of different numerical schemes for comparing amino acids, but it is important to remember that none can possibly capture the vast number of alternative environments that amino acids can find themselves in within proteins.
These pages get on the order of 30 000 hits per month (and rising; that's a hit about every minute on average), and yet to date I have only received a handful of E-mails. If you have comments or questions, please send them to me (russell@embl.de). I'm particularly interested in how the site might be improved according to what people are using it for.
Now this site has been running long enough, and been hit enough to do some stats. The table below shows how many times each page for each amino acid has been hit in the past 12 months (May 2003 - Apr 2004). I then compared the percentage of hits to the actual abundance of the amino acid. The ratio of these two gives some indication of the amino acids that people are most interested in. For instance, Trp, Cys, His and Arg are all big winners: hit many more times than one would expect compared to their abundances in proteins. Leu, Ile and Ser, in contrast, are the real boring losers. And my old boring favorite Ala is actually in the middle, so it just shows you that I don't know everything.
| AA | Hits | %Hits | %Abundance | Ratio |
| Trp | 1550 | 5.05 | 1.34 | 3.77 |
| Cys | 1905 | 6.21 | 1.76 | 3.53 |
| His | 1695 | 5.52 | 2.26 | 2.44 |
| Arg | 3093 | 10.08 | 5.20 | 1.94 |
| Met | 1102 | 3.59 | 2.32 | 1.55 |
| Tyr | 1525 | 4.97 | 3.25 | 1.53 |
| Gln | 1830 | 5.96 | 3.96 | 1.51 |
| Phe | 1616 | 5.27 | 4.12 | 1.28 |
| Asp | 1691 | 5.51 | 5.12 | 1.08 |
| Pro | 1598 | 5.21 | 5.00 | 1.04 |
| Ala | 2018 | 6.58 | 7.34 | 0.90 |
| Asn | 1103 | 3.59 | 4.57 | 0.79 |
| Lys | 1366 | 4.45 | 5.81 | 0.77 |
| Glu | 1471 | 4.79 | 6.22 | 0.77 |
| Gly | 1386 | 4.52 | 6.89 | 0.66 |
| Thr | 1147 | 3.74 | 5.85 | 0.64 |
| Val | 1225 | 3.99 | 6.48 | 0.62 |
| Ile | 1033 | 3.37 | 5.76 | 0.58 |
| Ser | 1206 | 3.93 | 7.38 | 0.53 |
| Leu | 1127 | 3.67 | 9.36 | 0.39 |
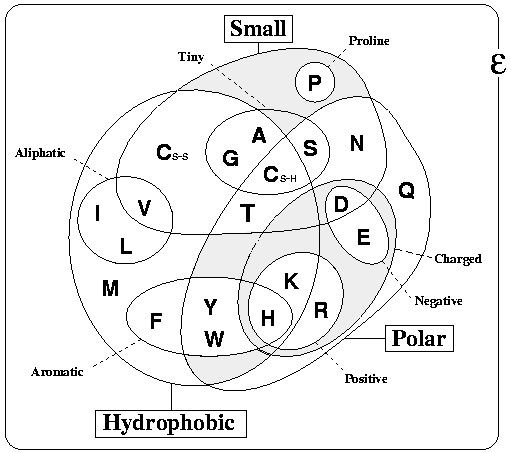
The figure below Venn diagram grouping amino acids according to their properties. This was adapted from Livingstone & Barton, CABIOS, 9, 745-756, 1993 (PubMed), and is just one of many classifications that are possible, but is probably that which most people would agree covers the most protein contexts.

(Now clickable!)
| Ala,A | Cys,C | Asp,D | Glu,E | Phe,F |
| Gly,G | His,H | Ile,I | Lys,K | Leu,L |
| Met,M | Asn,N | Pro,P | Gln,Q | Arg,R |
| Ser,S | Thr,T | Val,V | Trp,W | Tyr,Y |
| hydrophobic | aliphatic | aromatic | stacking |
| polar | charged | negative | positive |
| small | functional | C-beta branched |
Please also refer to the general explanation page to understand what the various sections on these pages mean, or how they are derived.
It is very difficult to put all amino acids of the same type into an invariant group. Remember that each amino acid is in a different protein environment, and its role in the protein is generally far more complicated than anything that could be defined by a set of groupings such as those given above. Although, the groupings are generally correct, it is critical to consider if your amino acid has a specific role in the protein, such as involvment in an active site, or such things as binding to a co-factor or a sugar, etc.
One critically important thing to consider is whether the amino acid of interest is conserved across known homologues. Invariant positions, or those showing conservation of certain residue properties (e.g. charge, hydrophobicity, etc.) are less likely to tolerate mutations than those where the protein family permits mutations to a great variety of amino acids. When doing this, also remember the important distintinction between orthologues, which are proteins with the same function in the different species, and paralogues, which are the result of duplications within a species (i.e. homologues probably performing different functions). A position conserved across orthologues can be involved in may aspects of structure & function, including specificity, whereas one conserved across paralogues can only be assumed to be involved in structure, and more general functional features such as catalytic mechanism.
There is no easy way to discern orthologues and paralogues automatically, particularly when considering big evolutionary distances (e.g. human and yeast), but two proteins from different species are often considered to be orthologous if they detect each other as the best match during (say) Blast searches, and if there are no other proteins with scores in the same range.
There are some general trends with regard to amino acids that prefer to be in functional centres, but bear in mind that these are averages and the importance of a particular type of amino acid to function varies greatly with the functional context.
Something specifically important to notice are the two positions of Cysteine (C-SS and C-SH). These denote the two oxidation states of Cysteines. C-SS denotes those cysteines that are involved in disulphide bonds (i.e. connected to each other). Note that these bonds are extremely rare in intracellular proteins, so if your protein is intracellular, the cysteines are more likely to be free (C-SH). This means that Cysteines have quite different properties in the extra and intra cellular environments. More information can be found here.