 |
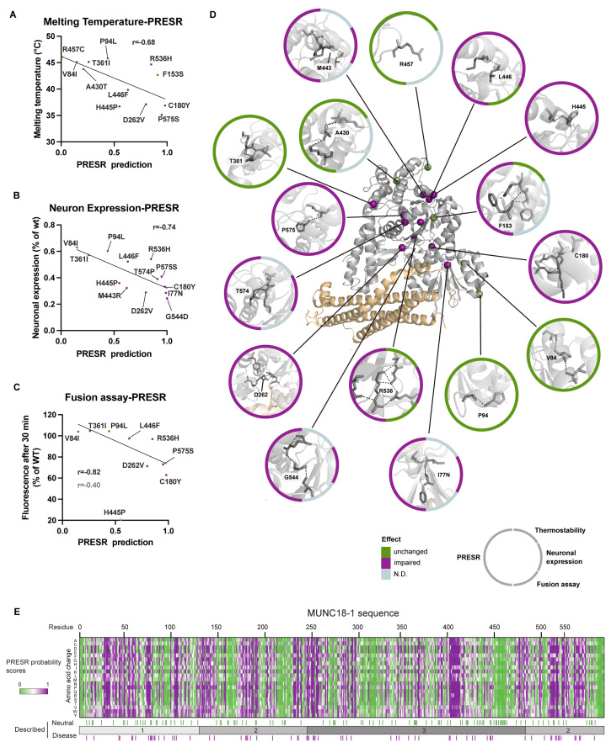
Characterizing variants in STXBP1: Display of variants predicted as disease (purple) or neutral (green) in STXBP1 and associated biophysical measurements |
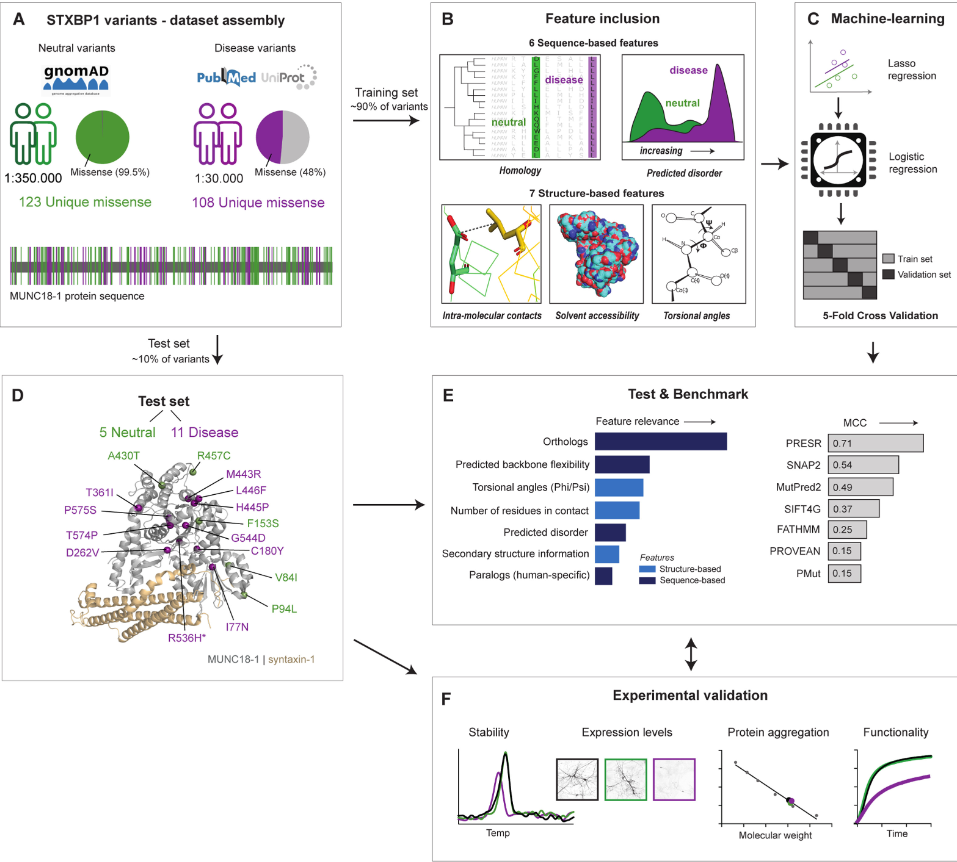 |
Characterizing variants in STXBP1: Machine learning schema for the STXBP1 predictor |
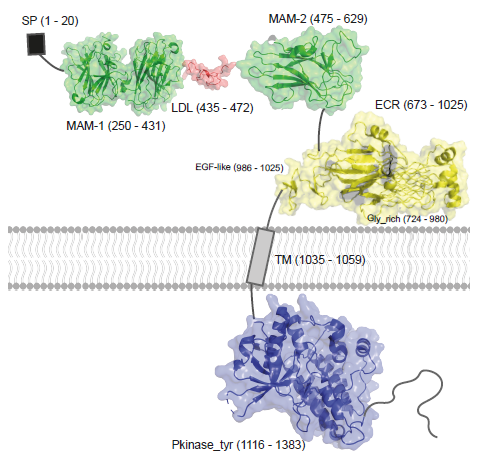 |
ALK variants in sarcomas: Predicted structure of full-length ALK |
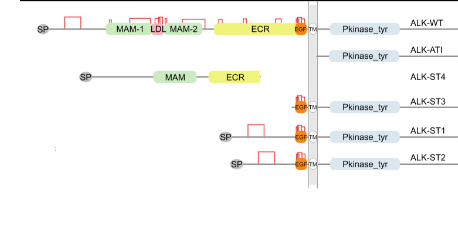 |
ALK variants in sarcomas: Truncation variants of ALK in sarcomas |
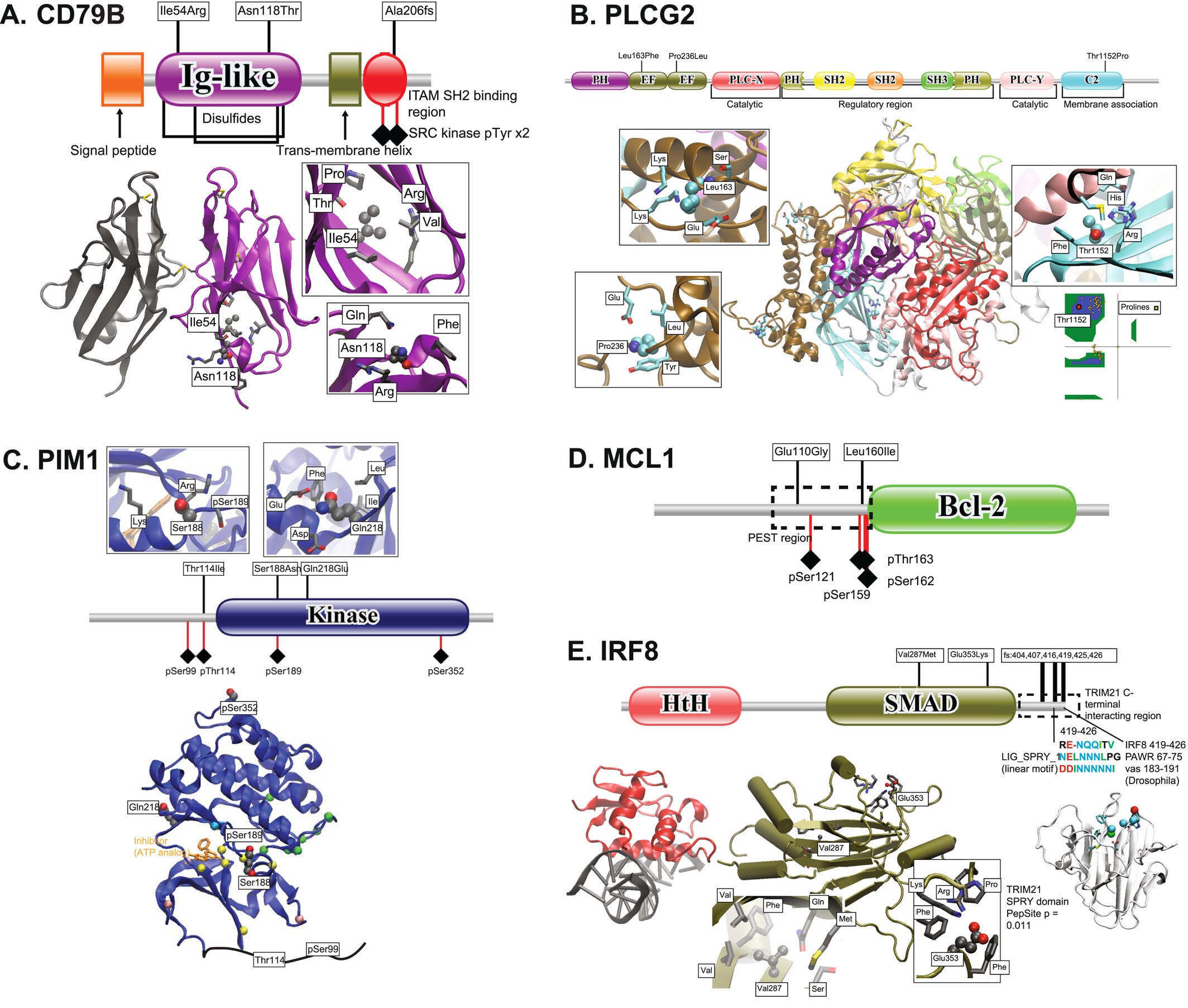 |
Genomic landscape of follicular lymphoma: Various structure/domain images suggesting how variants in FL affect protein function. |
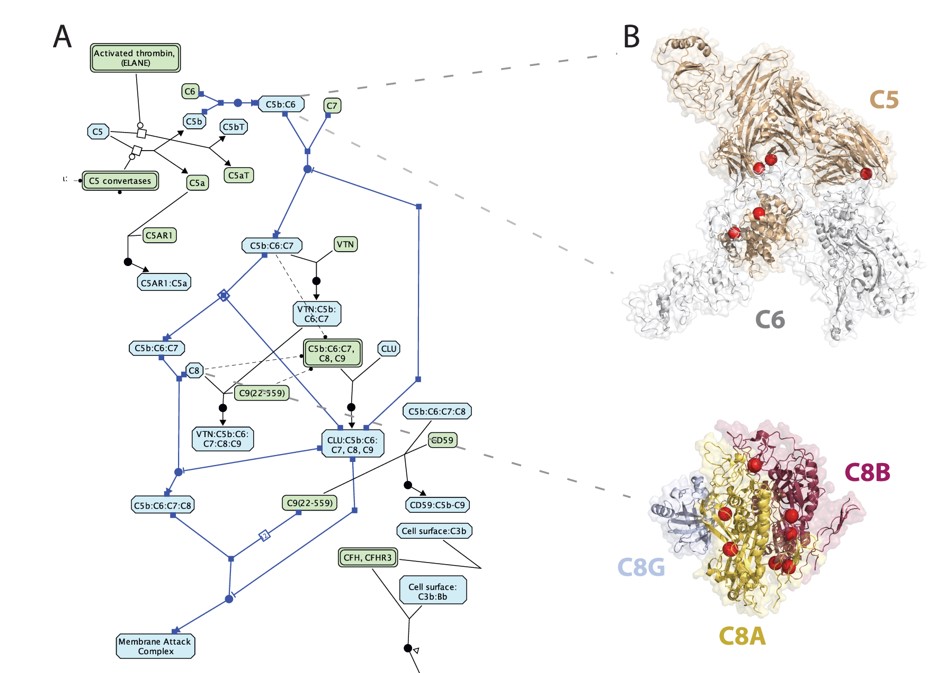 |
Integrating protein interfaces and Reactome to study cancer variants: Reactome view of the complement cascade pathway (left); non-synonymous cancer variants (spheres) at interface structures identified via Mechismo (right). |
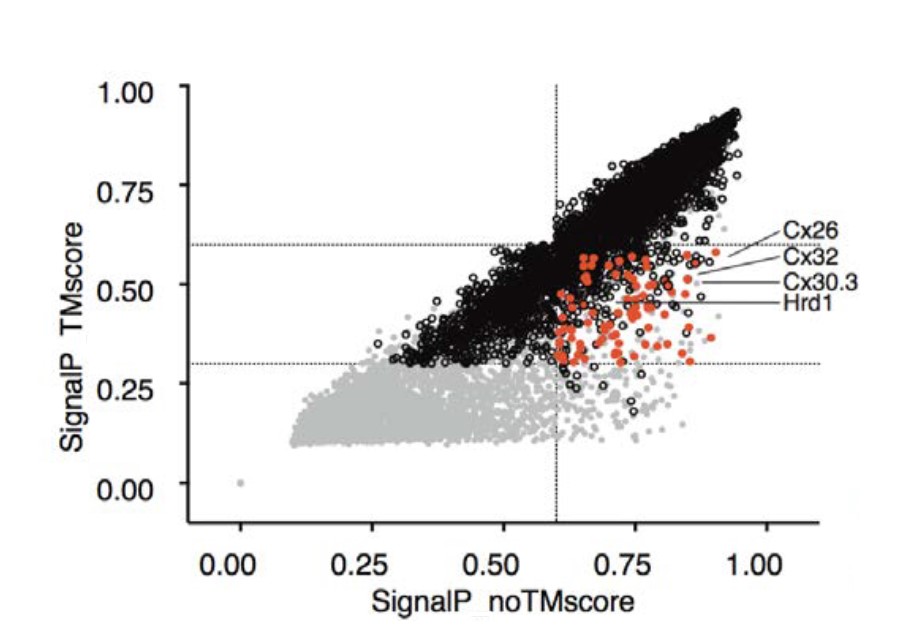 |
Signal Peptidase as a QC enzyme for membrane proteins: Plot showing potential cleavage sites adjacent to TM segments |
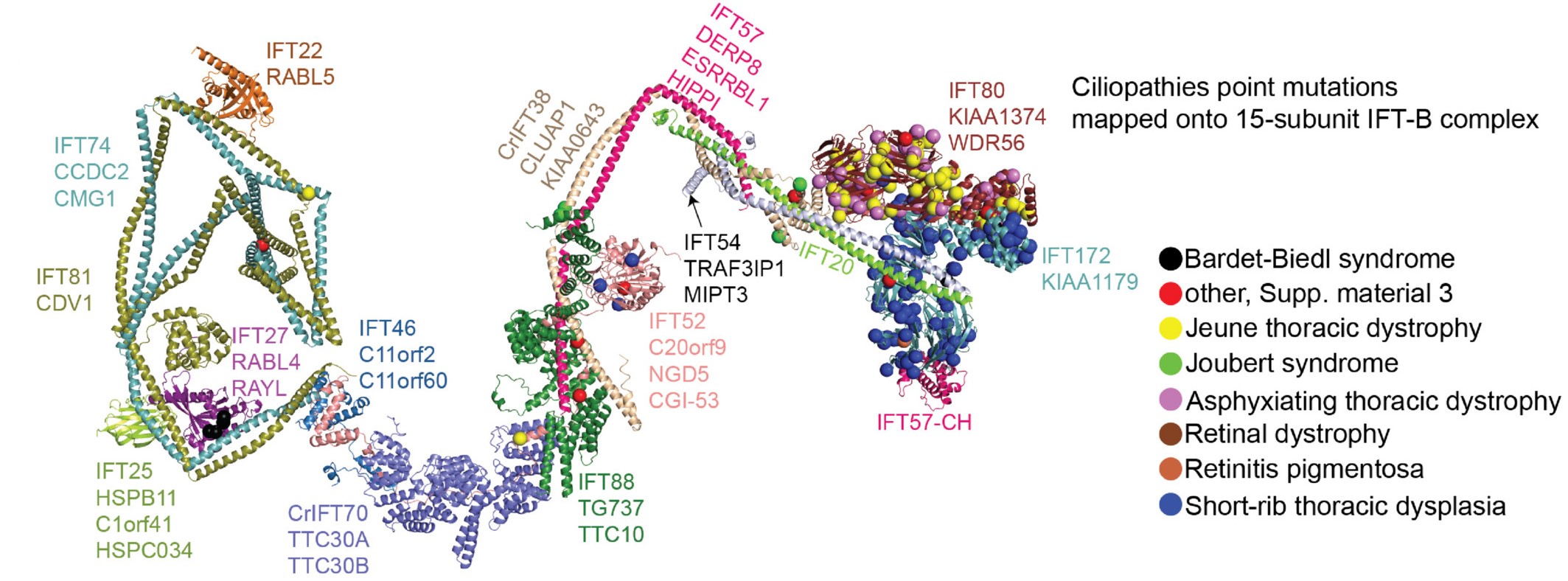 |
IFT-B 3D model with disease variants mapped: Ciliopathy mutations shown on the model of IFT-B (made with the group of Esben Lorenzen, Aarhus) |
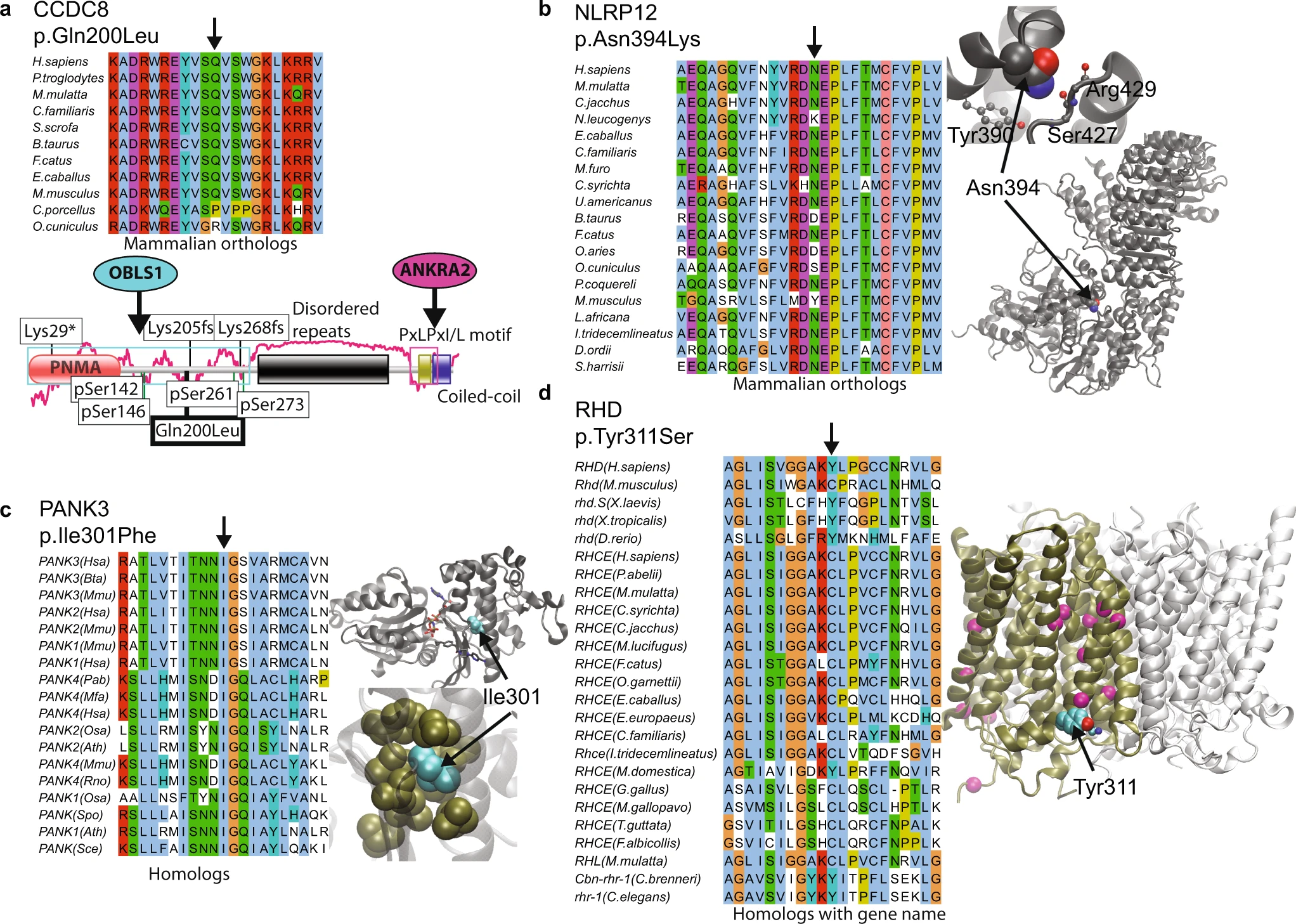 |
Never-homozygous natural variants as disease candidates: candidates of exclusively heterozygous variants where there is compelling functional reasons for possible disease association |
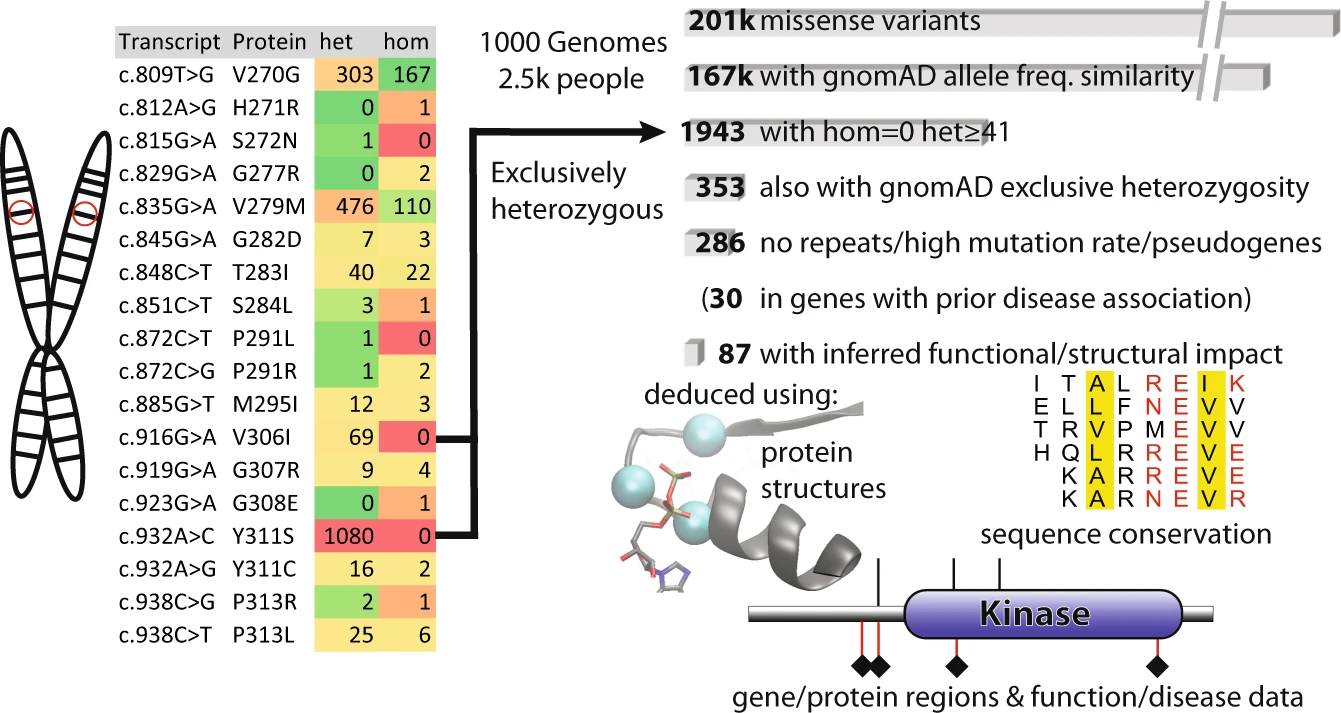 |
Never-homozygous natural variants as disease candidates: overview of how exclusively heterozygous variants were defined and scored |
 |
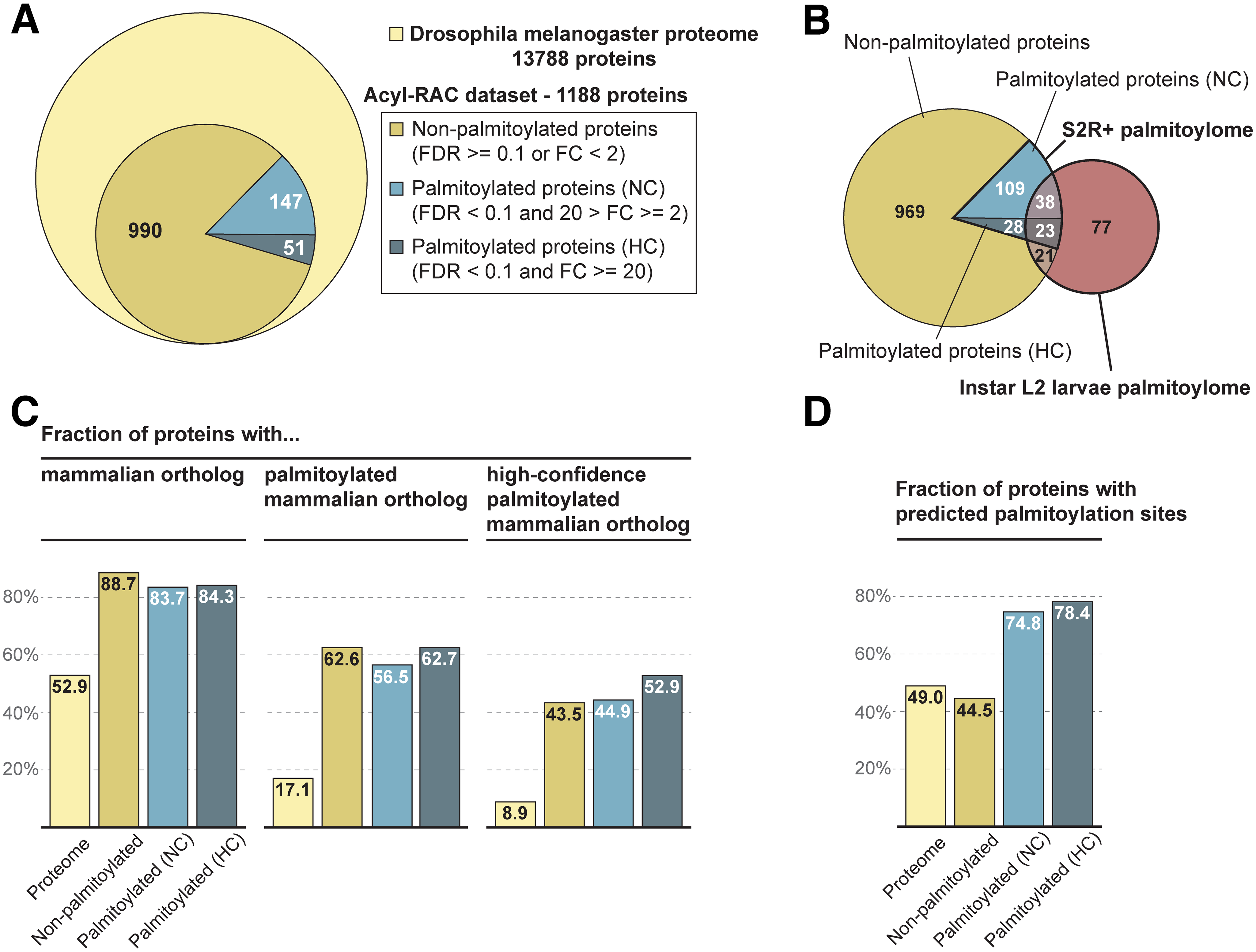
The Drosophila palmitoylome: overview of the dataset and comparison with earlier studies |
 |
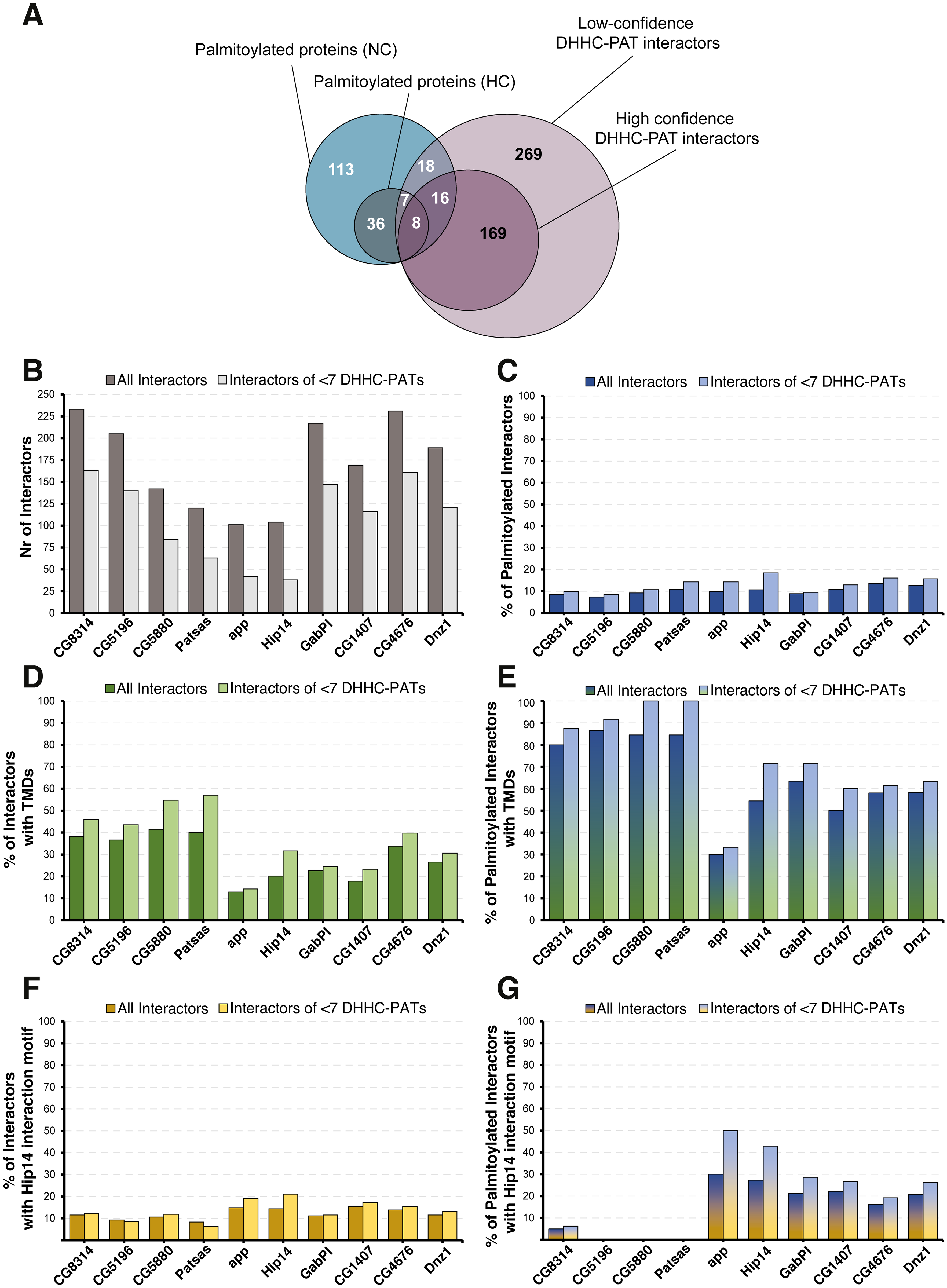
The Drosophila palmitoylome: overlap between palmitoylated proteins and interactions with DHHC-PAT enzymes |
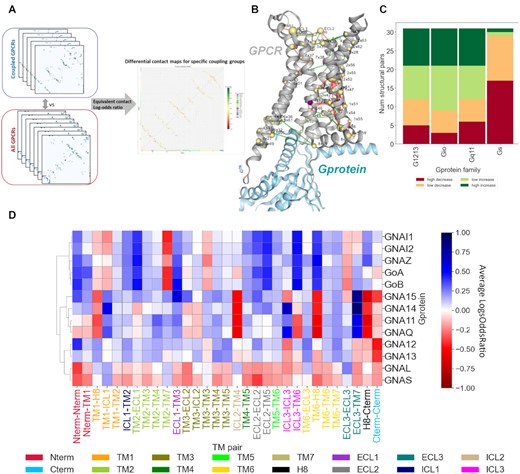 |
PRECOGx: predicting GPCR signaling: scheme and results for how differential contacts resolves GPCR transducer interactions |
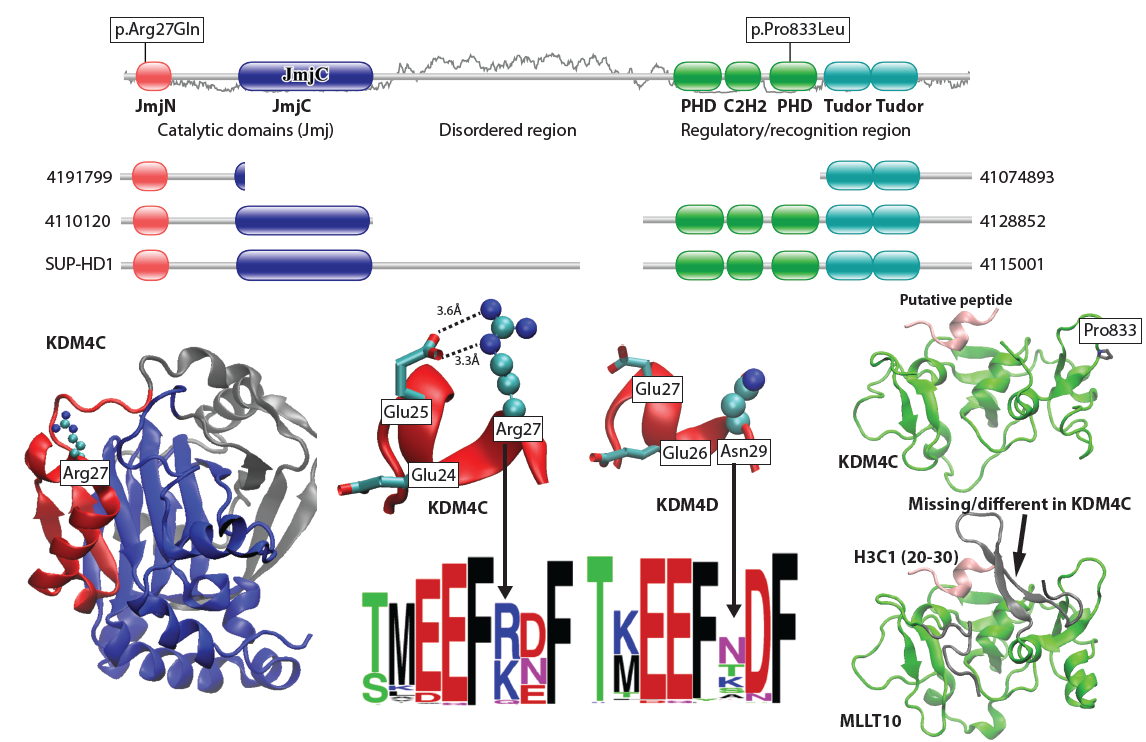 |
KDM4C variants in Lymphomas: our mechanistic analyses of various KDM4C truncation (top) and missense variants and how they might influence protein function. |
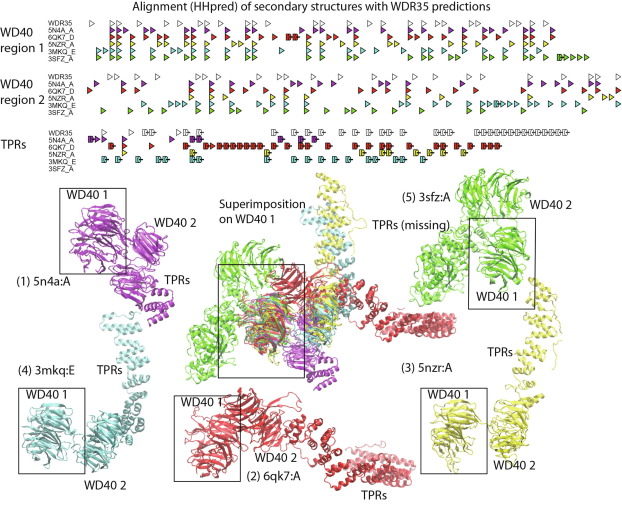 |
Alphafold2 for genetic variant interpretation: an illustration of the problems with predicting certain, repeat-rich, human proteins |
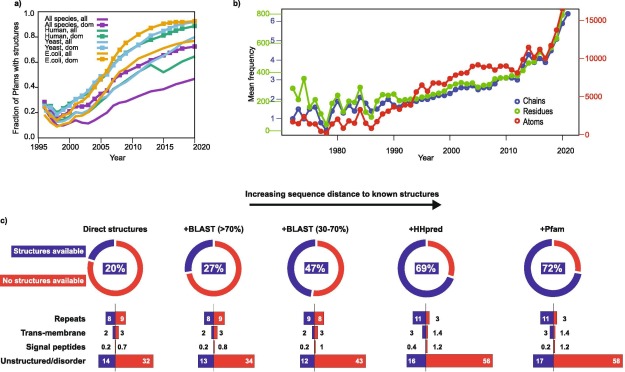 |
Alphafold2 for genetic variant interpretation: data on the current size, and structural coverage of human proteins as of mid 2021 |
 |
Mechnetor: new protein mechanism explorer: examples of different interaction mechanisms displayed in Mechnetor |
 |
Mechnetor: new protein mechanism explorer: examples of different interaction mechanisms displayed in Mechnetor |
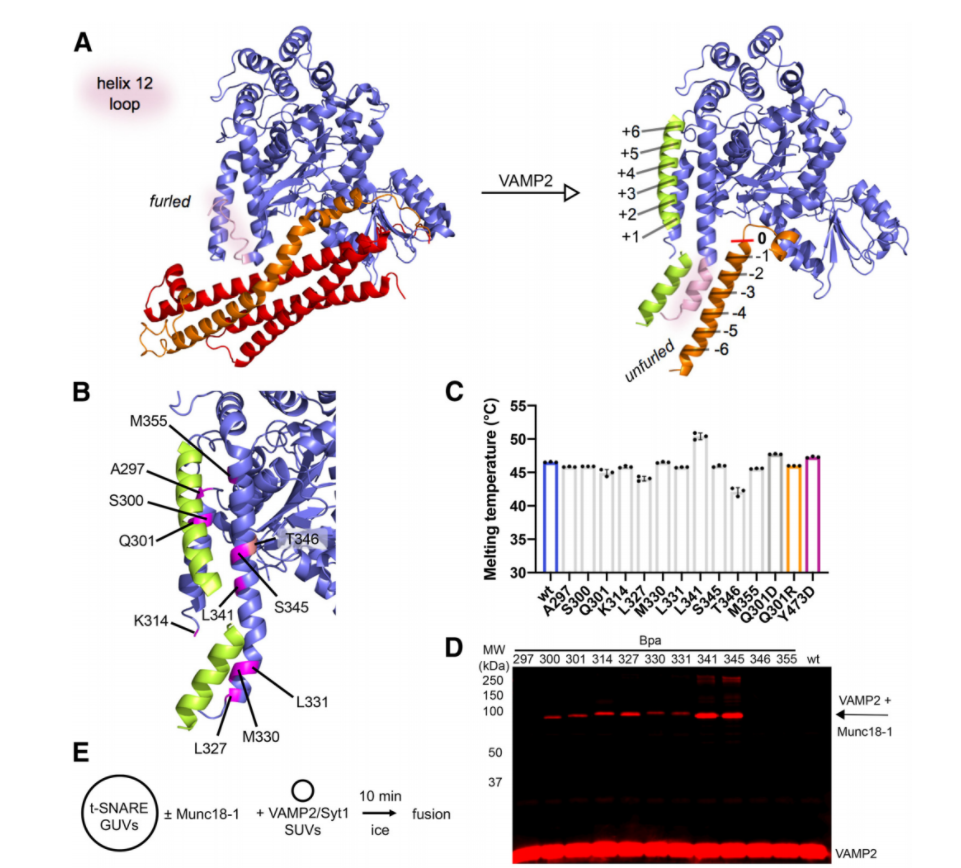 |
Dissecting the role of Munc18 in SNARE: A model of the interaction between Munc18 and the VAMP2 SNARE motif. |
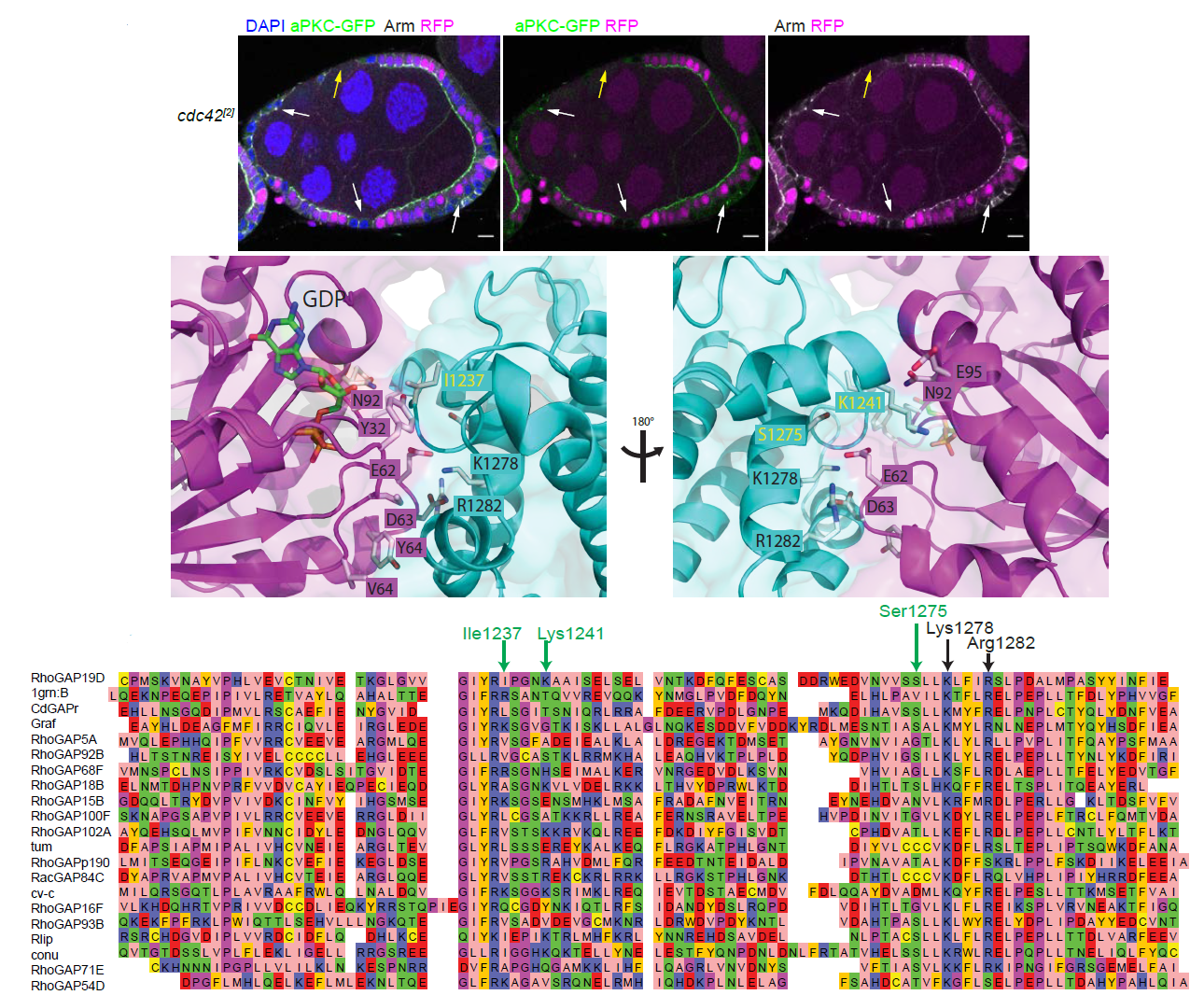 |
Predicting and validating RhoGAP functions: A model of how one particular Drosophila GAP molecule might bind to Cdc42. |
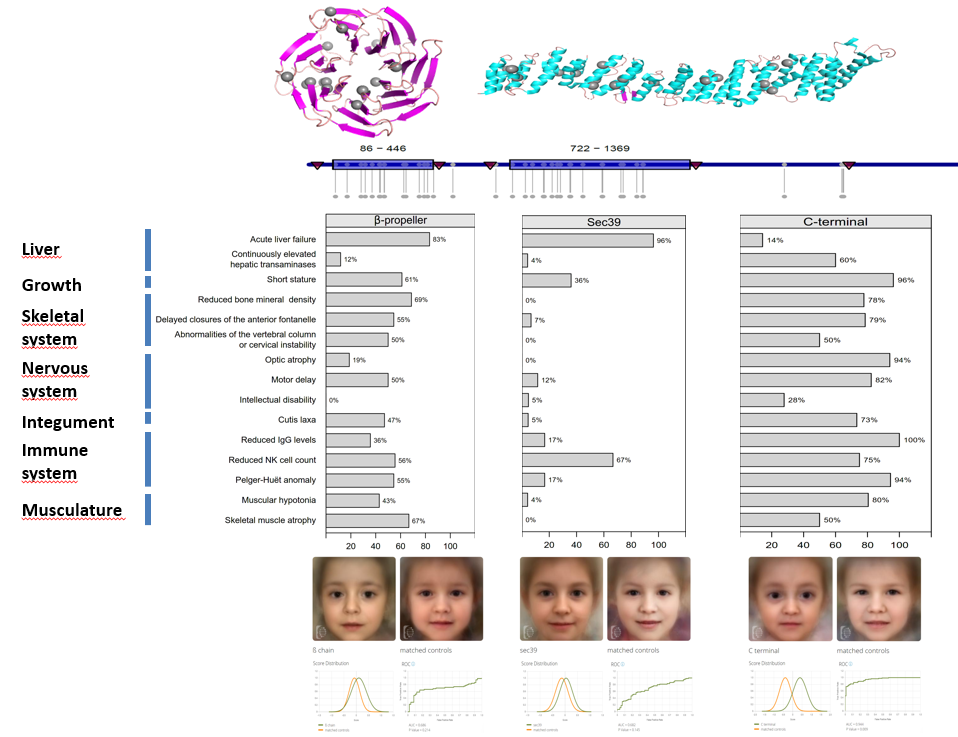 |
Genetics variants in NBAS-associated diseases: Location of mutations in NBAS and how their location on the encoded protein correlates with different disease phenotypes. |
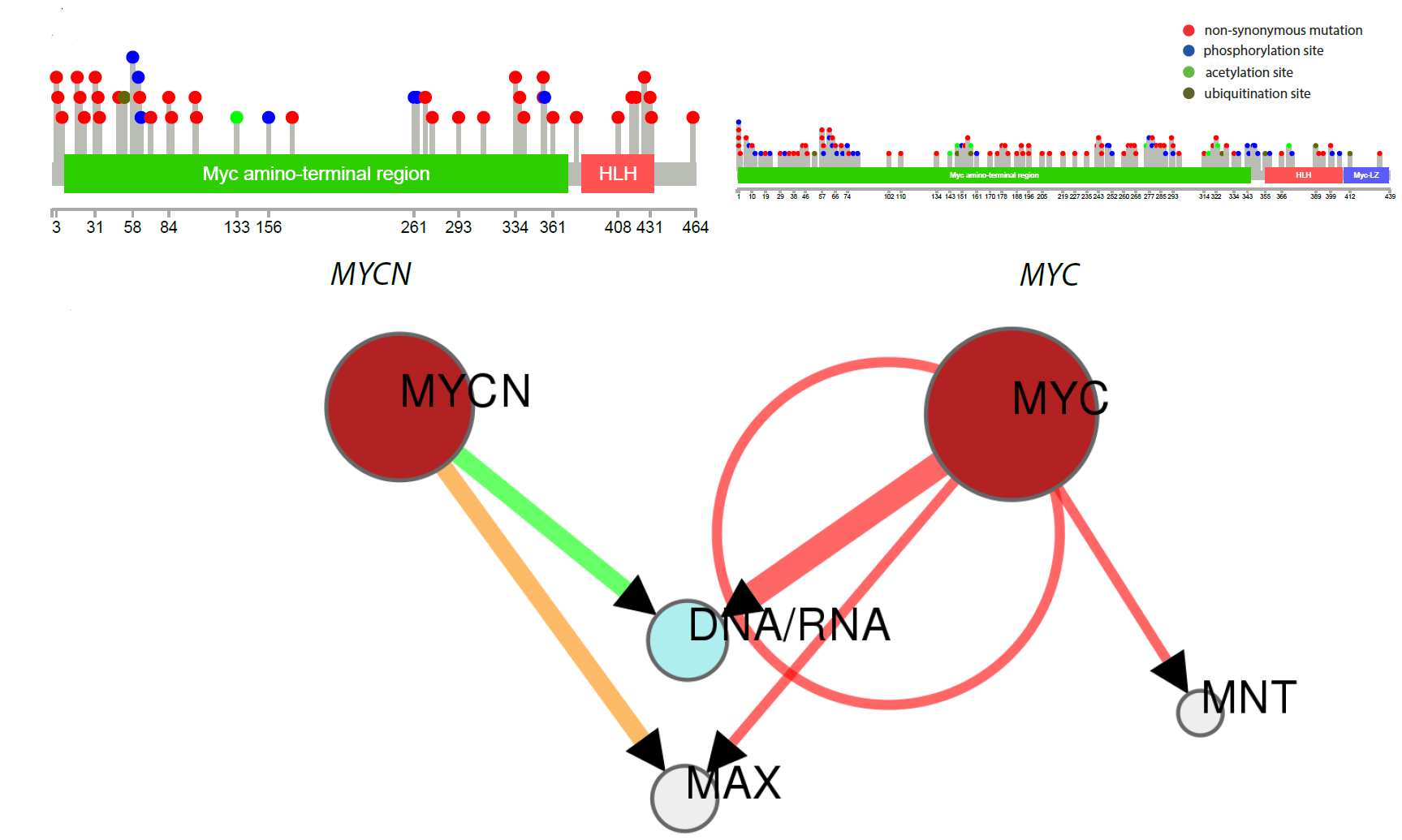 |
N-MYC variants replacing MYC in Burkitt lymphoma : Location of mutations in N-MYC and MYC in patients with Burkitt lymphoma, and how they are predicted to alter protein/DNA interactions. |
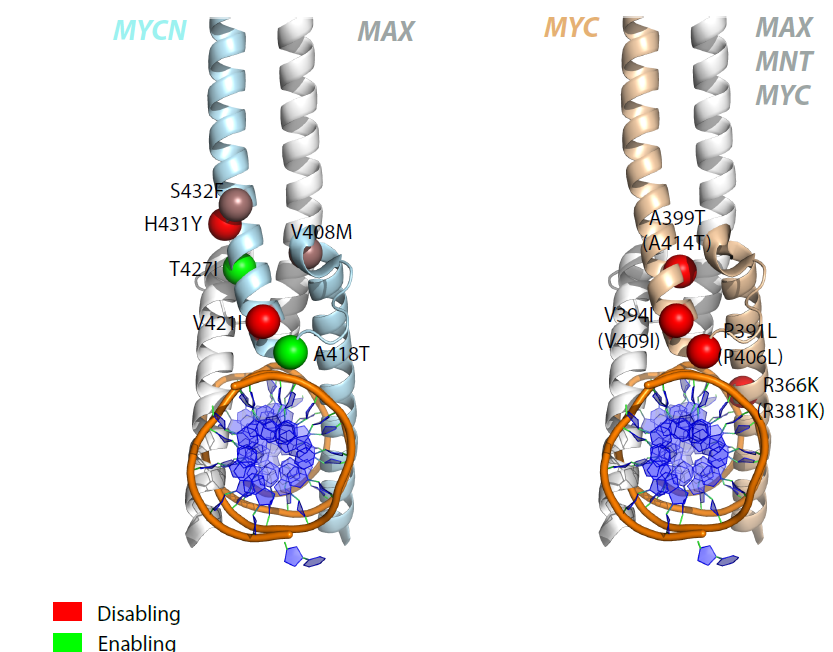 |
N-MYC variants replacing MYC in Burkitt lymphoma : Location of mutations in N-MYC and MYC structures highlighting possible functional differences. |
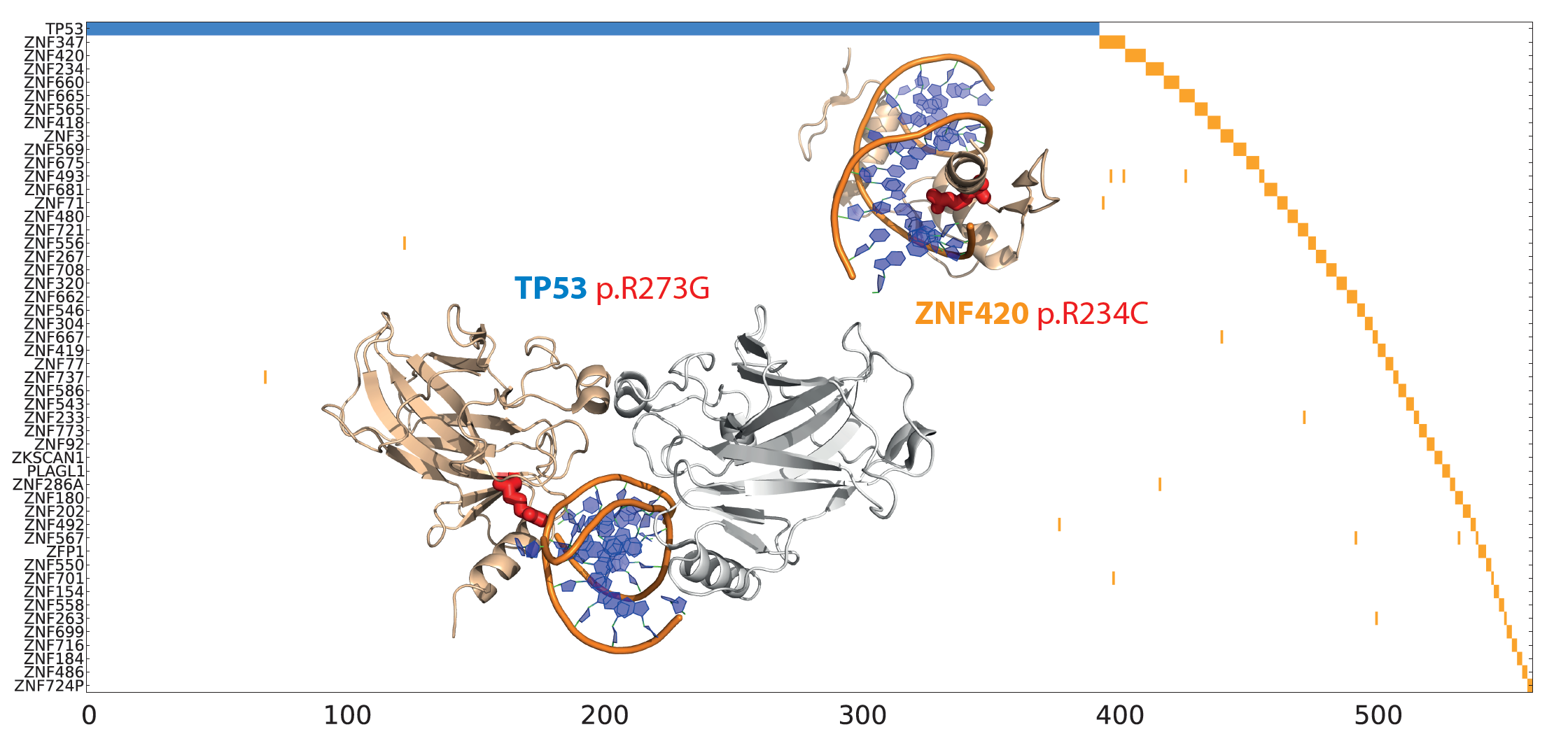 |
Multi-gene oncodrivers: Mutual exclusivity (in terms of tumor mutations) of a key functional mutation in P53 with similar DNA binding mutations in multiple Zinc fingers. |
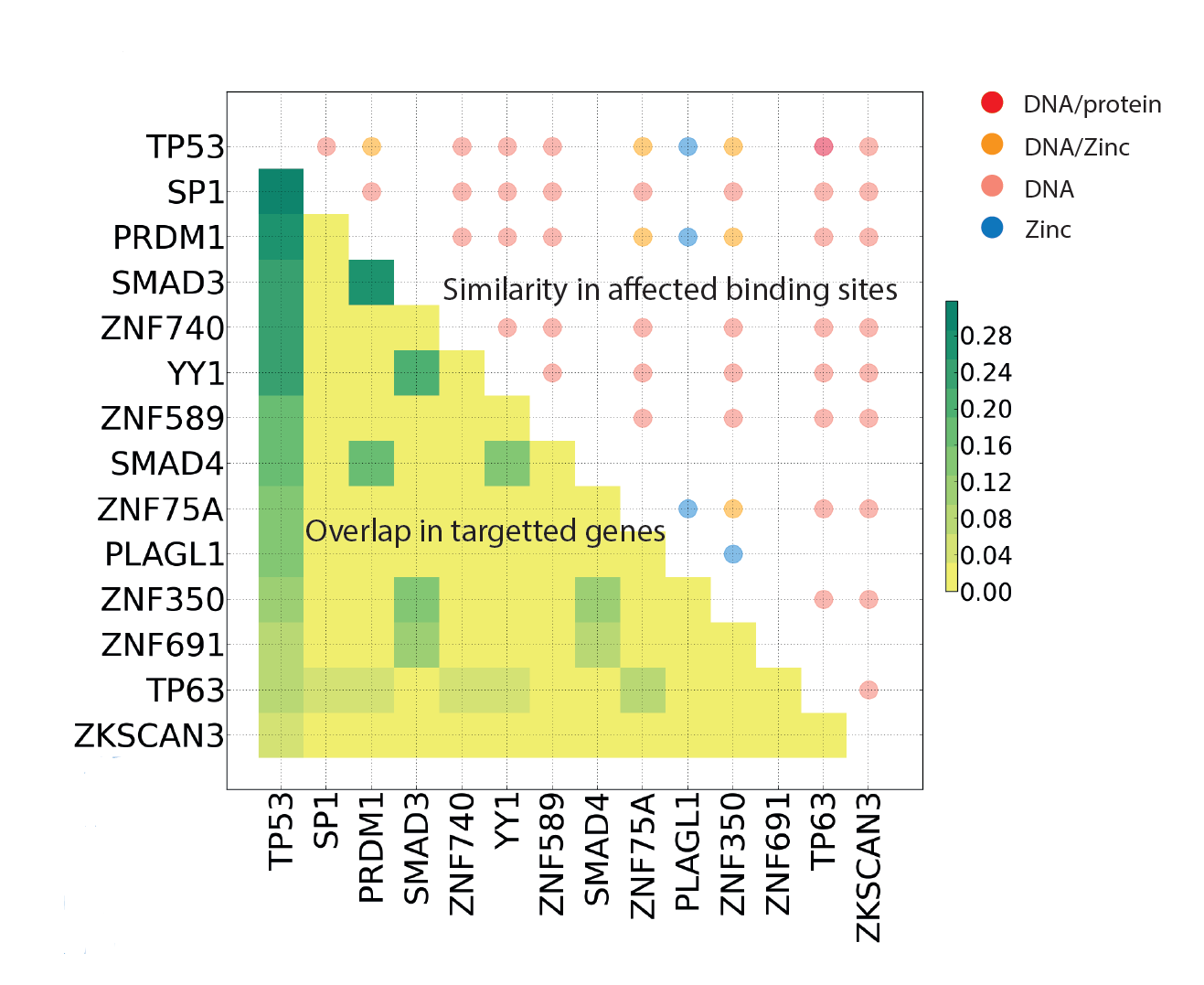 |
Multi-gene oncodrivers: Similarity in targeted genes (ChIP-Seq) and function (Mechismo) of P53 and mutually exclusive Zinc fingers. |
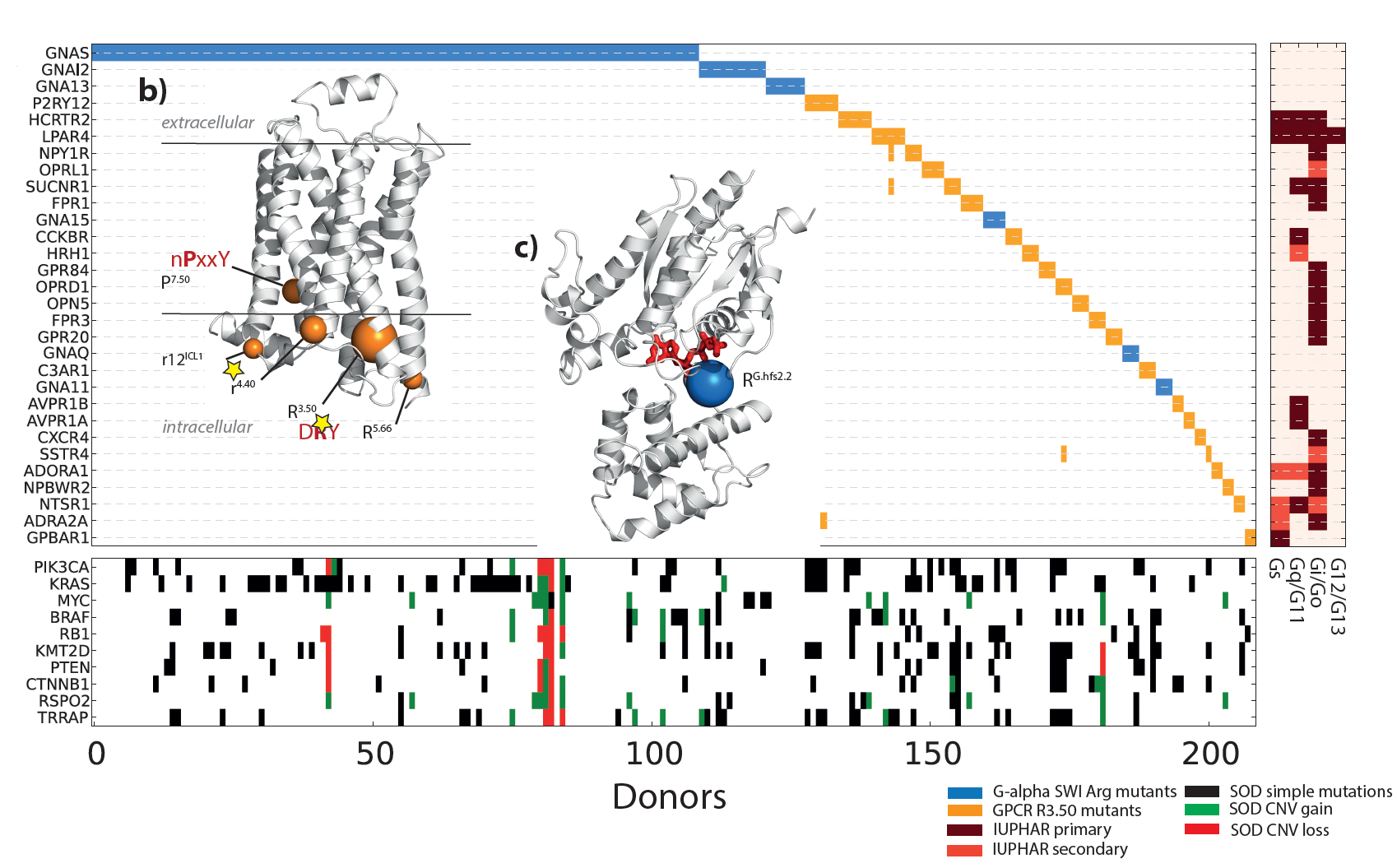 |
Multi-gene oncodrivers: Mutual exclusivity (in terms of tumor mutations) of a key functional mutation in G-proteins and another in multiple GPCRs. |
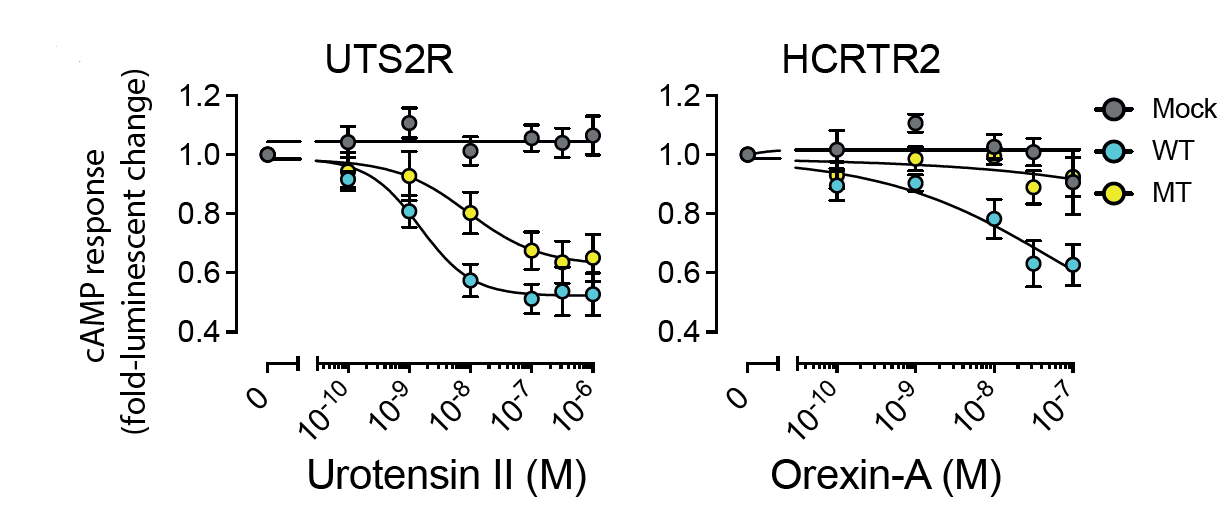 |
Multi-gene oncodrivers: Increase in cAMP production by mutations in the DRY motif in two GPCRs (also caused by GNAS mutations). |
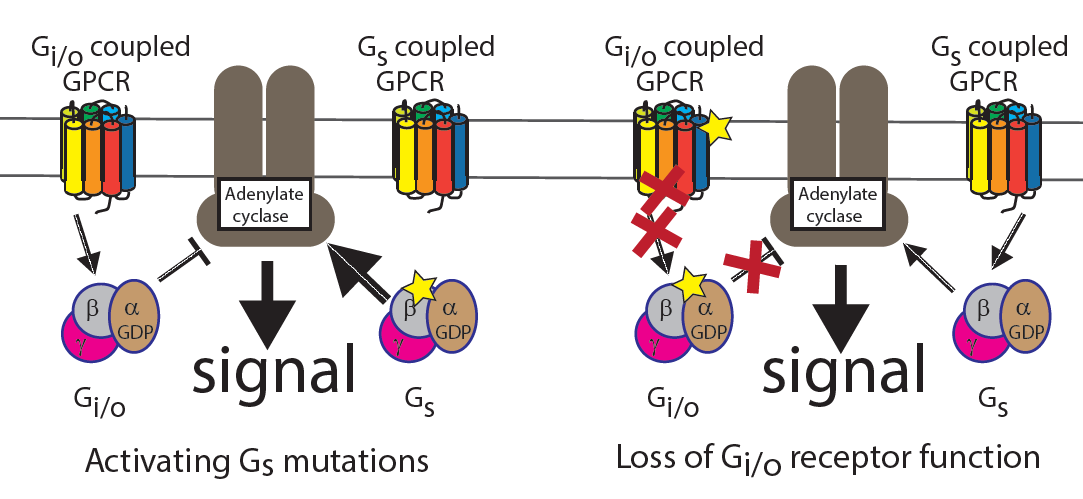 |
Multi-gene oncodrivers: Model of how activation of G-protein Gs and deactivation of Gi/o GPCRs leads to the same effect. |
 |
Illuminating GPRC/G-protein coupling: Cover illustration (sadly not selected). Created by Thomas Splettstößer at SciStyle. |
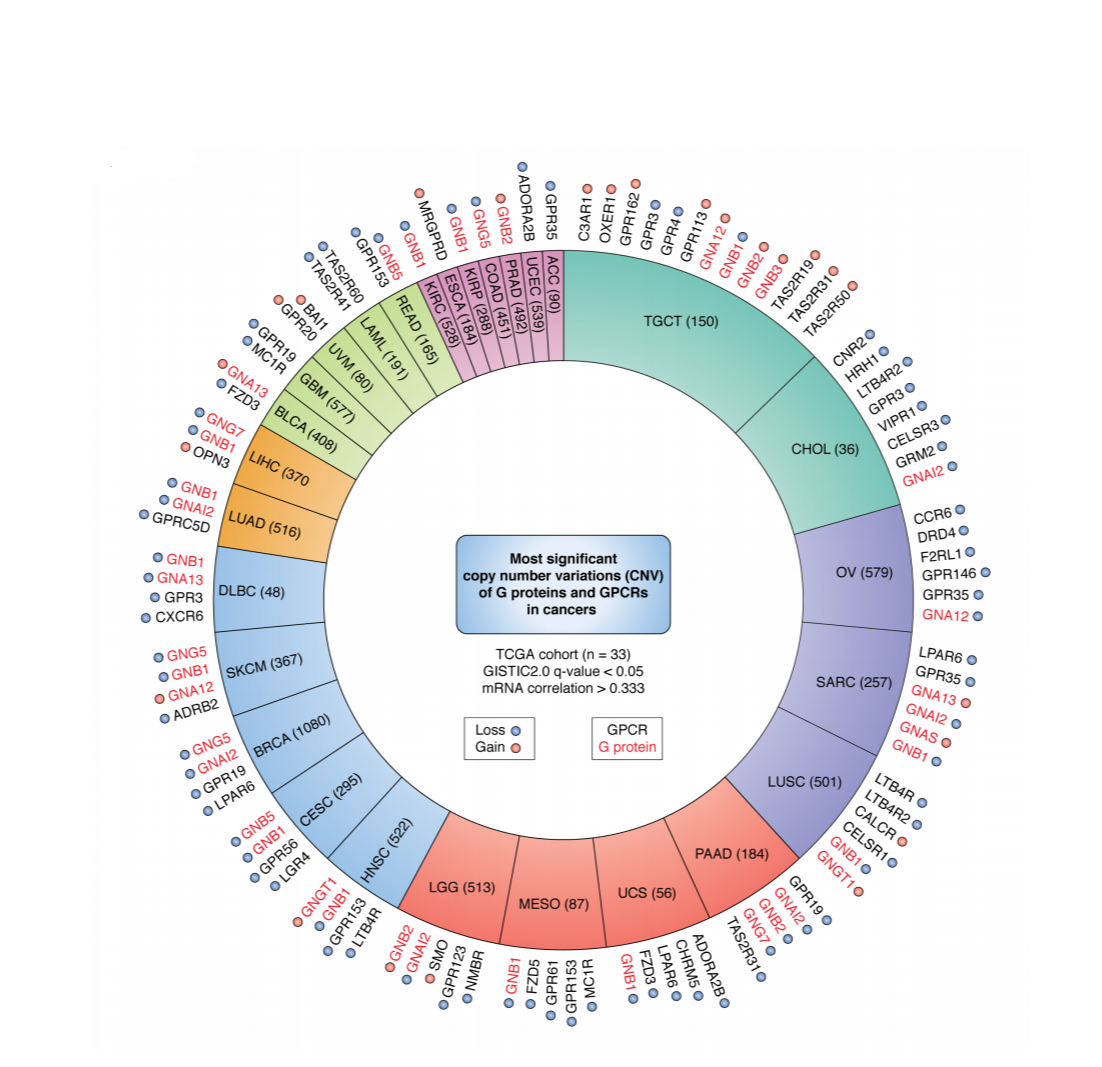 |
The GPCRome in Cancer: Copy number variants in GPCRs and G-proteins across multiple cancer (somatic) genomes |
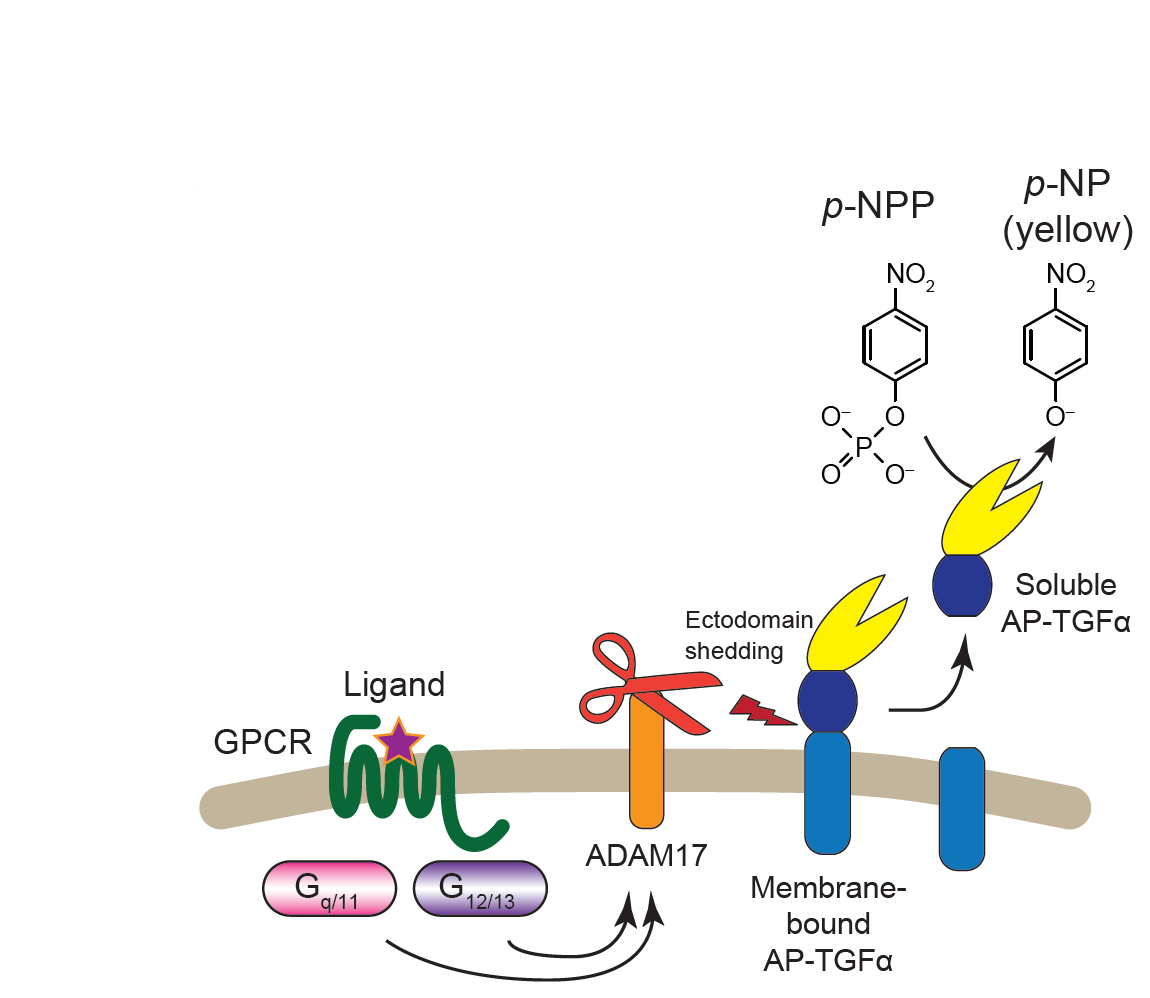 |
Illuminating GPRC/G-protein coupling: Schema the TGFalpha shedding assay used for high-throughput assessment of GPCR/G-protein coupling |
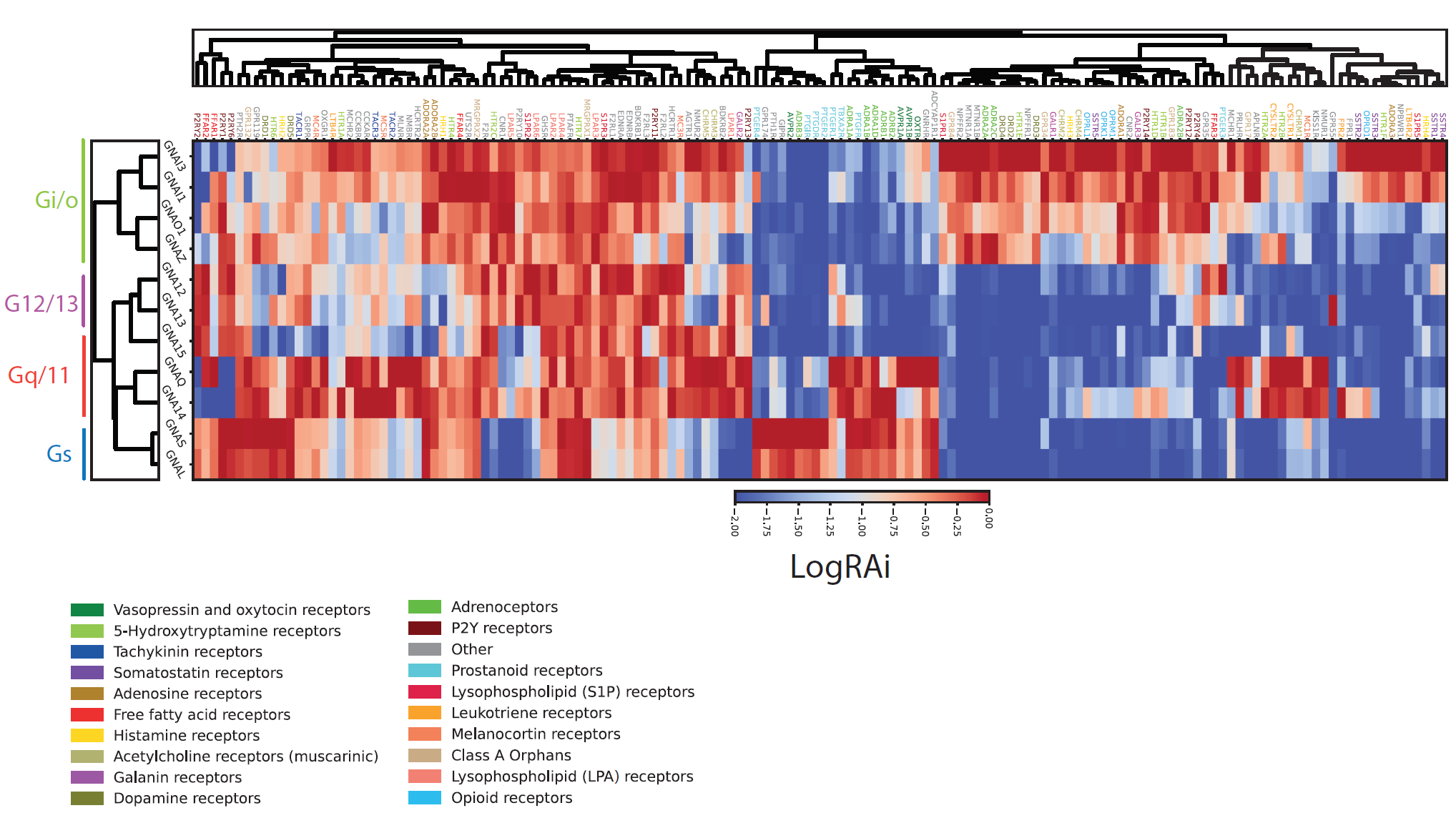 |
Illuminating GPRC/G-protein coupling: Coupling profiles for 148 representative human GPCRs and 11 G-proteins |
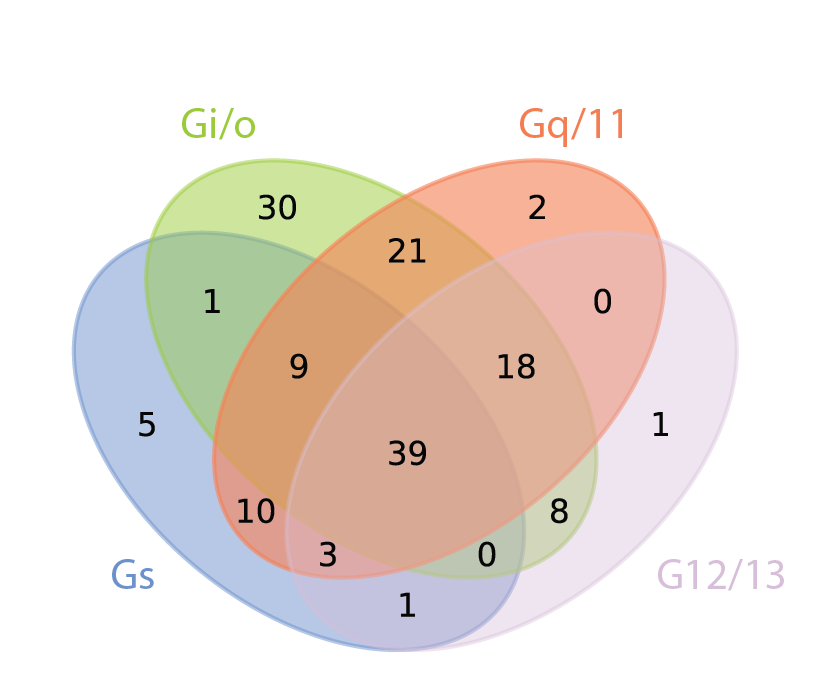 |
Illuminating GPRC/G-protein coupling: How GPCRs couple to one or more G-proteins. Counts indicate how many receptors couple to each of the four main G-protein subfamilies |
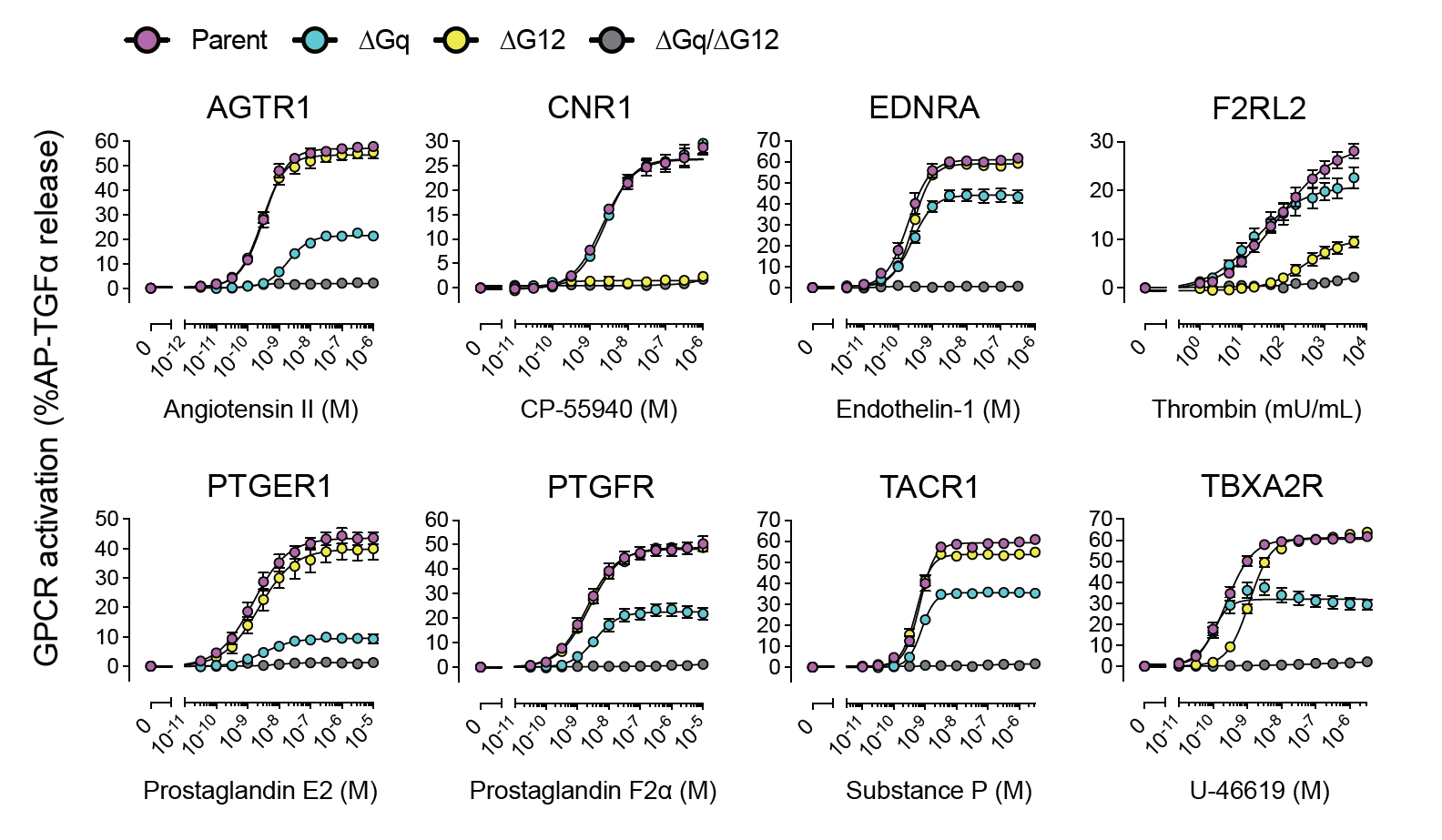 |
Illuminating GPRC/G-protein coupling: Curves illustrating measurement of G-protein binding for different recpetors via the TGF-alpha shedding assay |
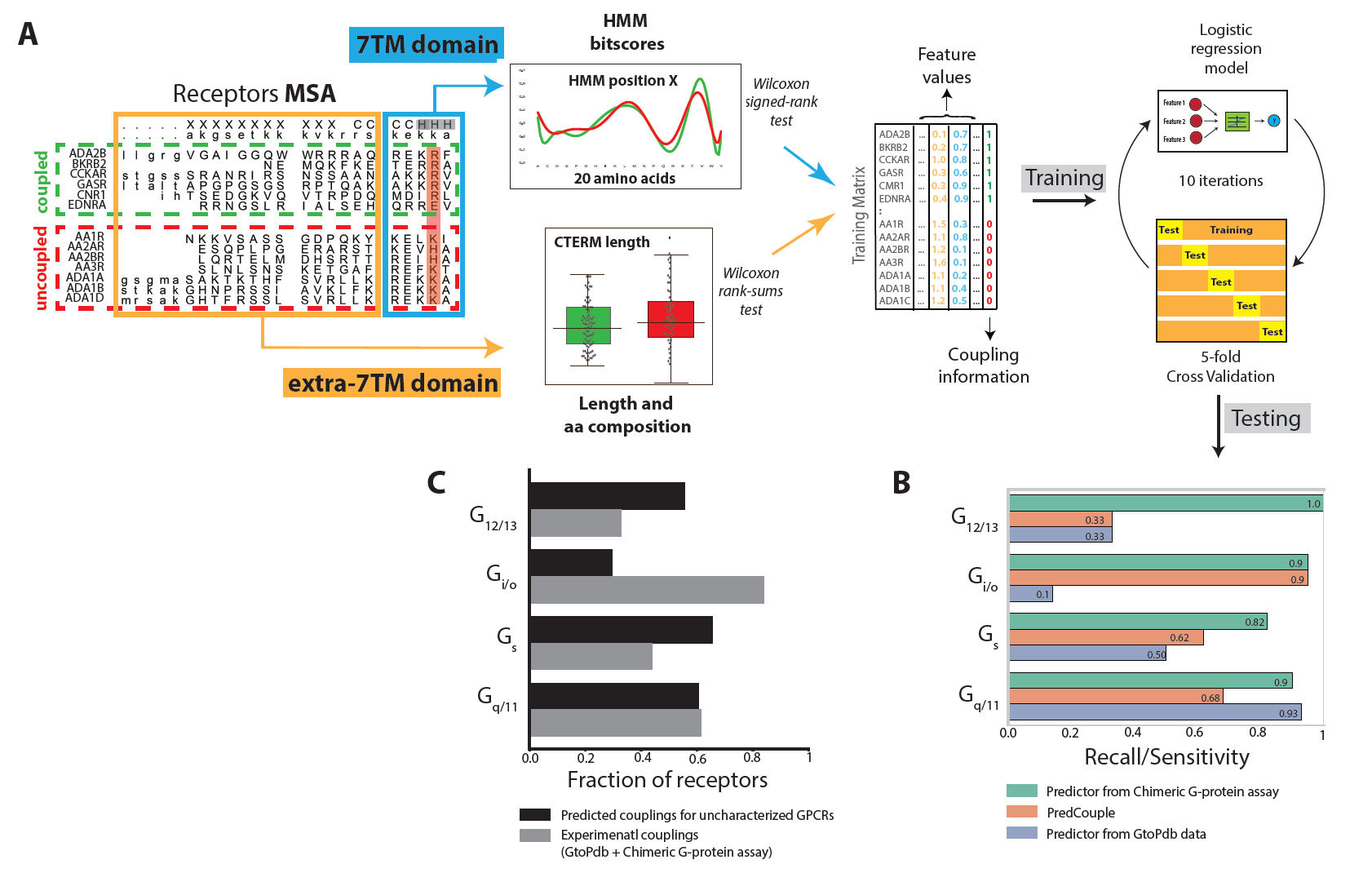 |
Illuminating GPRC/G-protein coupling: Schema for training and testing the logistic regression algorithm for predicting coupling by Machine Learning |
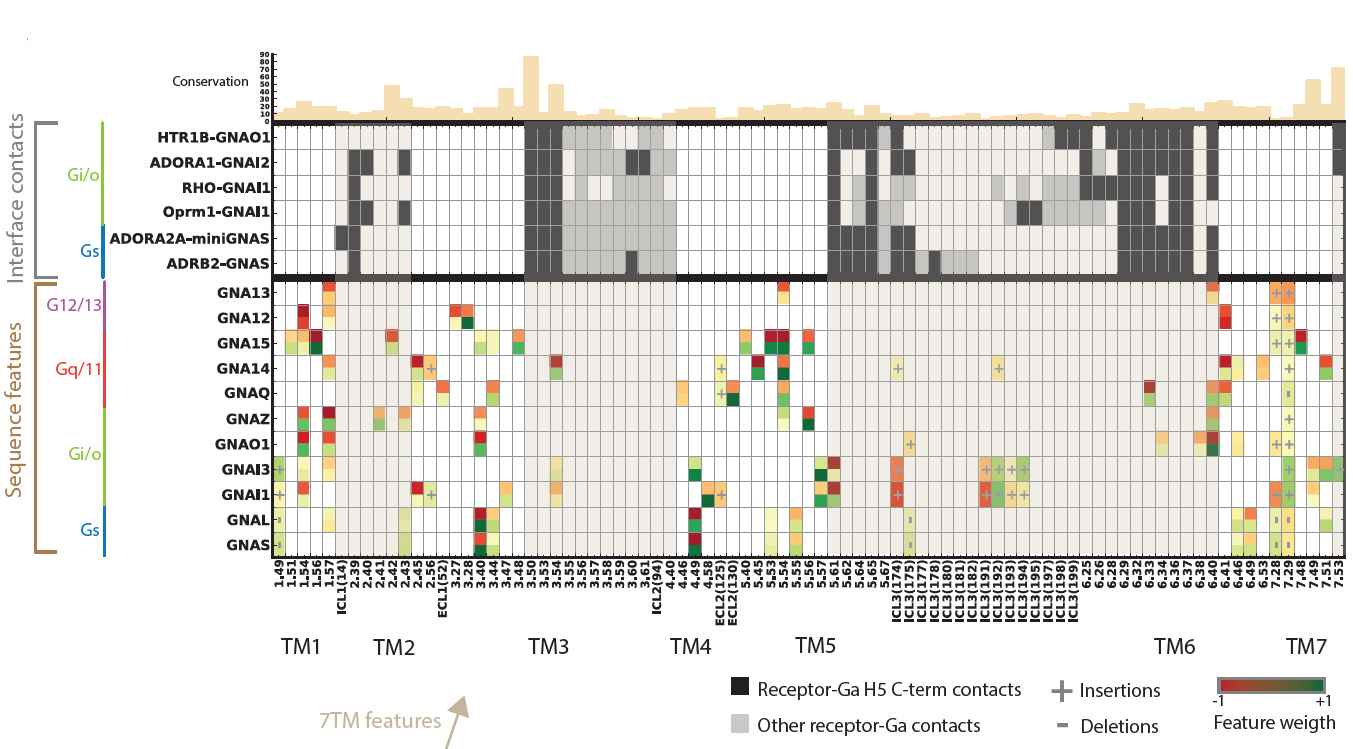 |
Illuminating GPRC/G-protein coupling: Positions in the GPCR structure identified as being predictive of coupling to each G-protein |
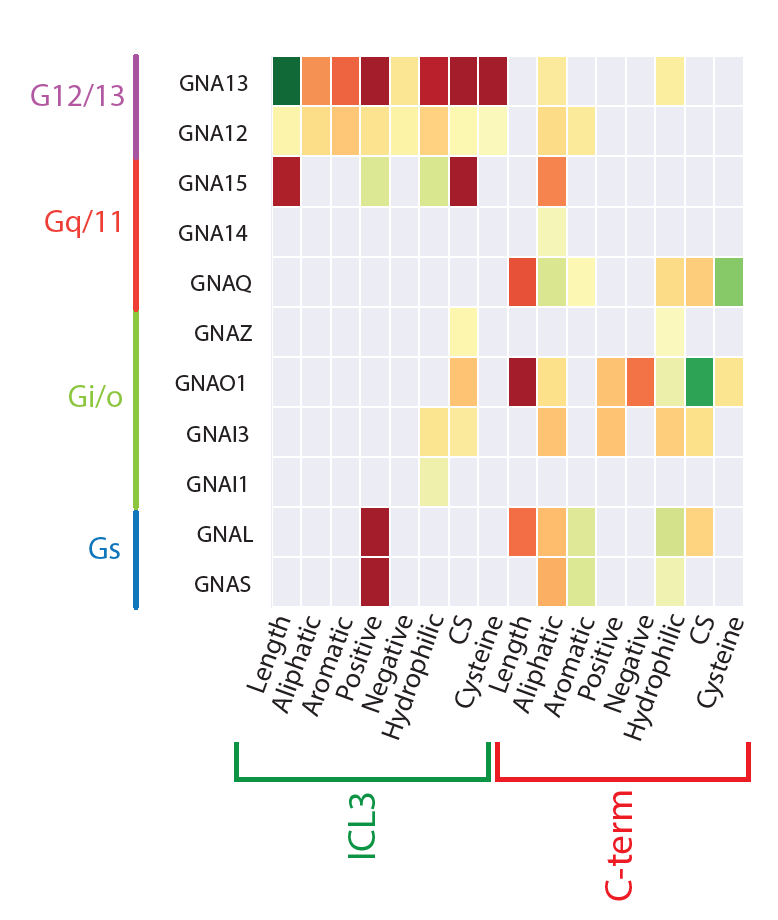 |
Illuminating GPRC/G-protein coupling: Sequence properties in the third intracellular loop (ICL3) and the C-terminus predictive of coupling to each G-protein |
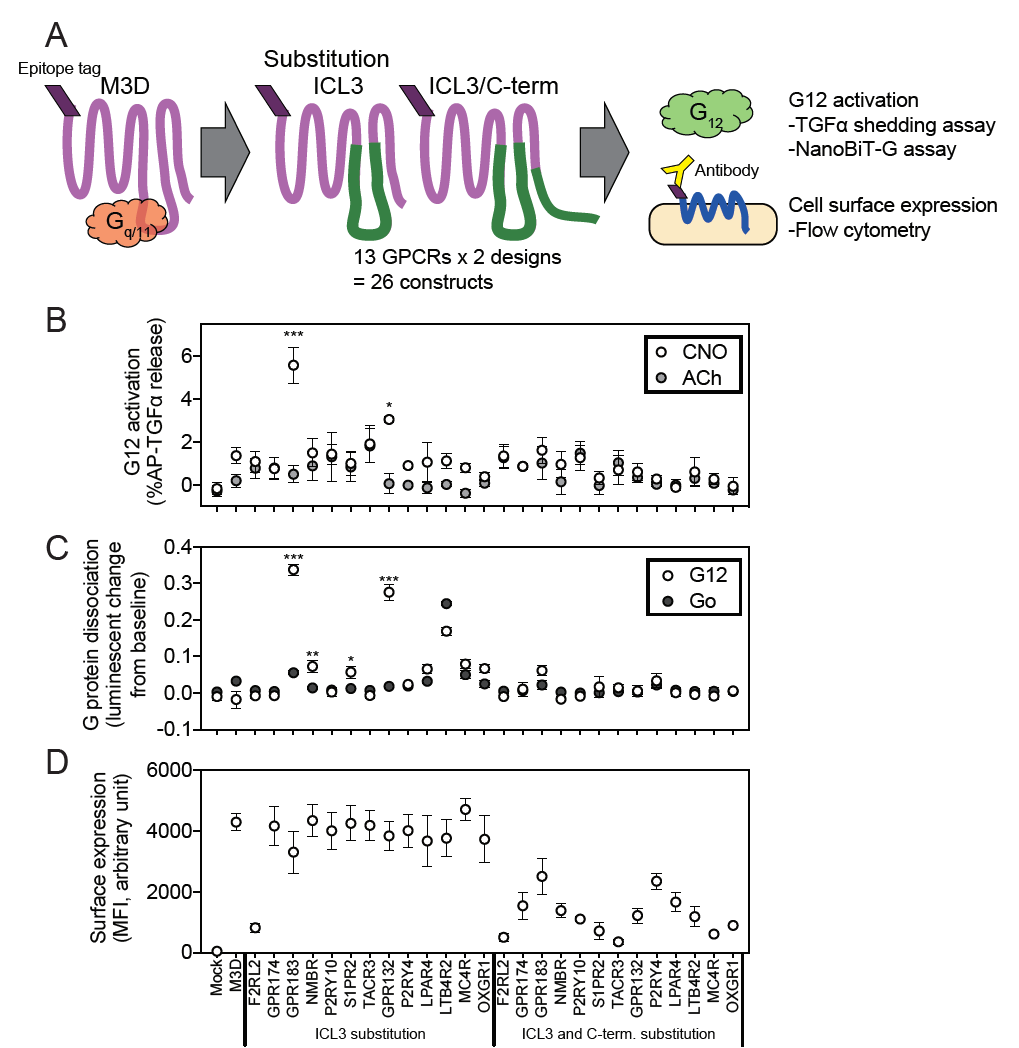 |
Illuminating GPRC/G-protein coupling: Design and testing of a novel chemogenetic tool (DREADD receptor) specifically signalling through G-alphpa 12 |
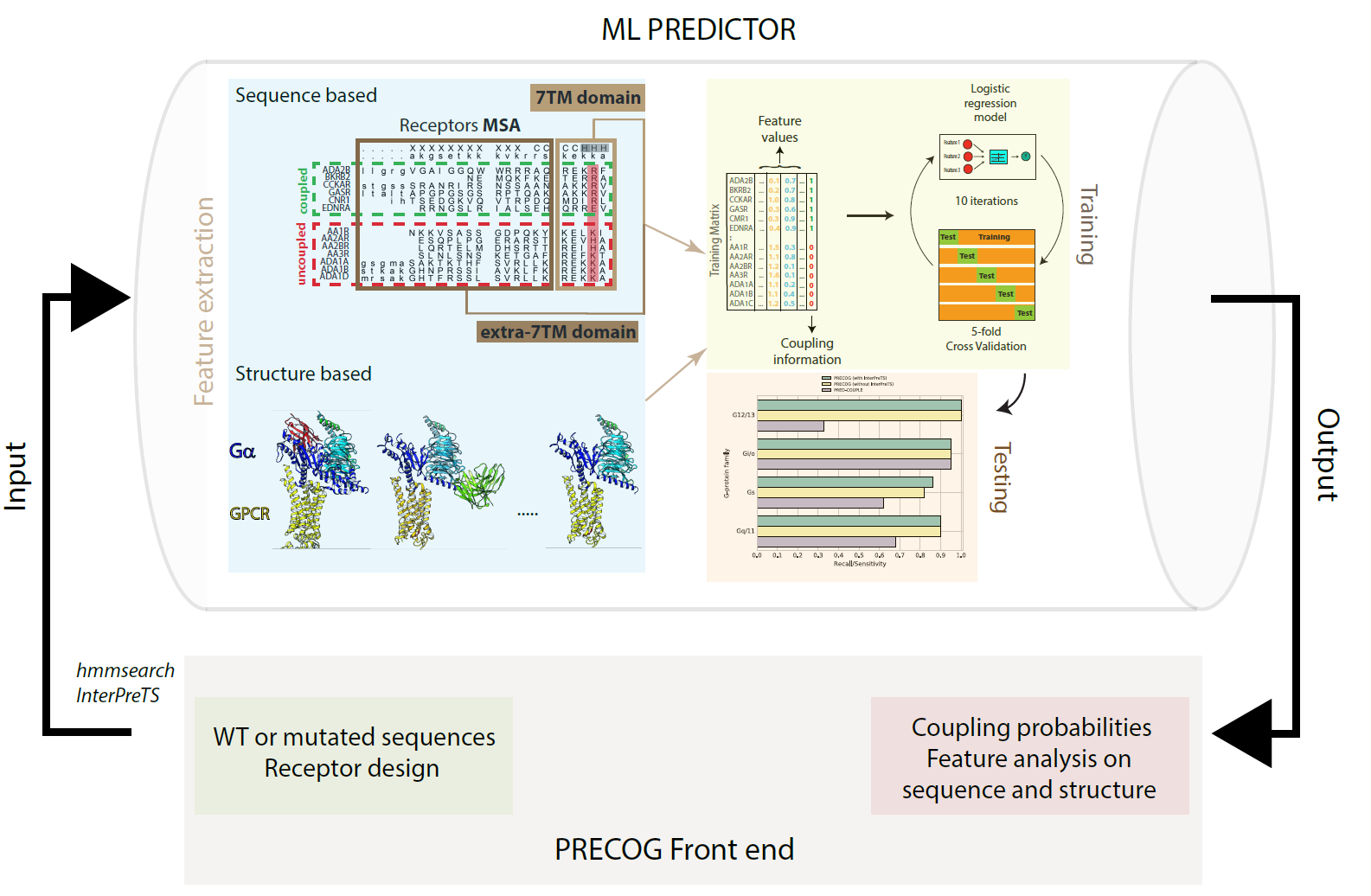 |
Predicting GPCR/G-protein coupling: Schema of our GPCR/G-protein coupling predictor |
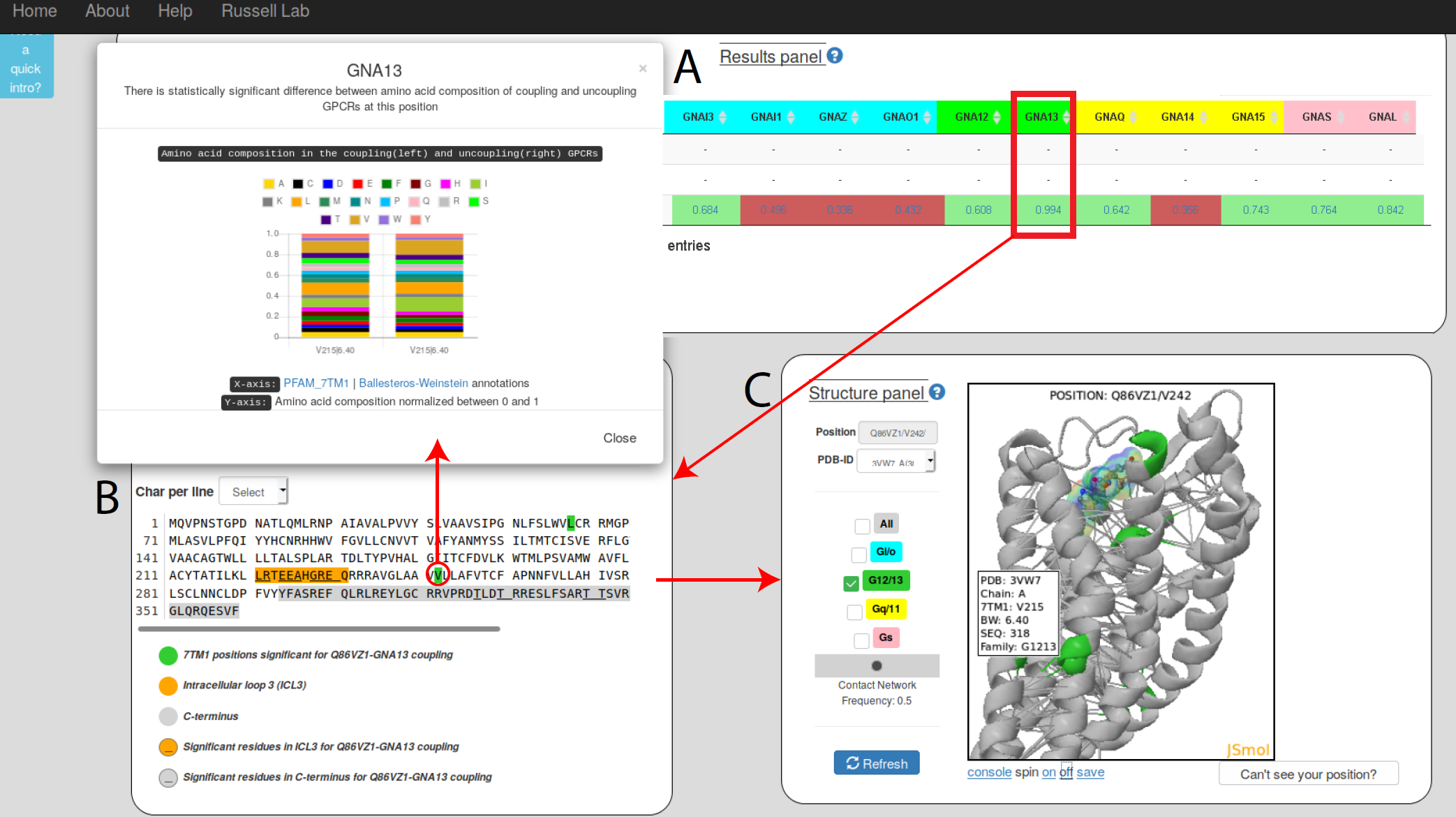 |
Predicting GPCR/G-protein coupling: Example of GPCR/G-protein output showing a mutation predicted to affect coupling |
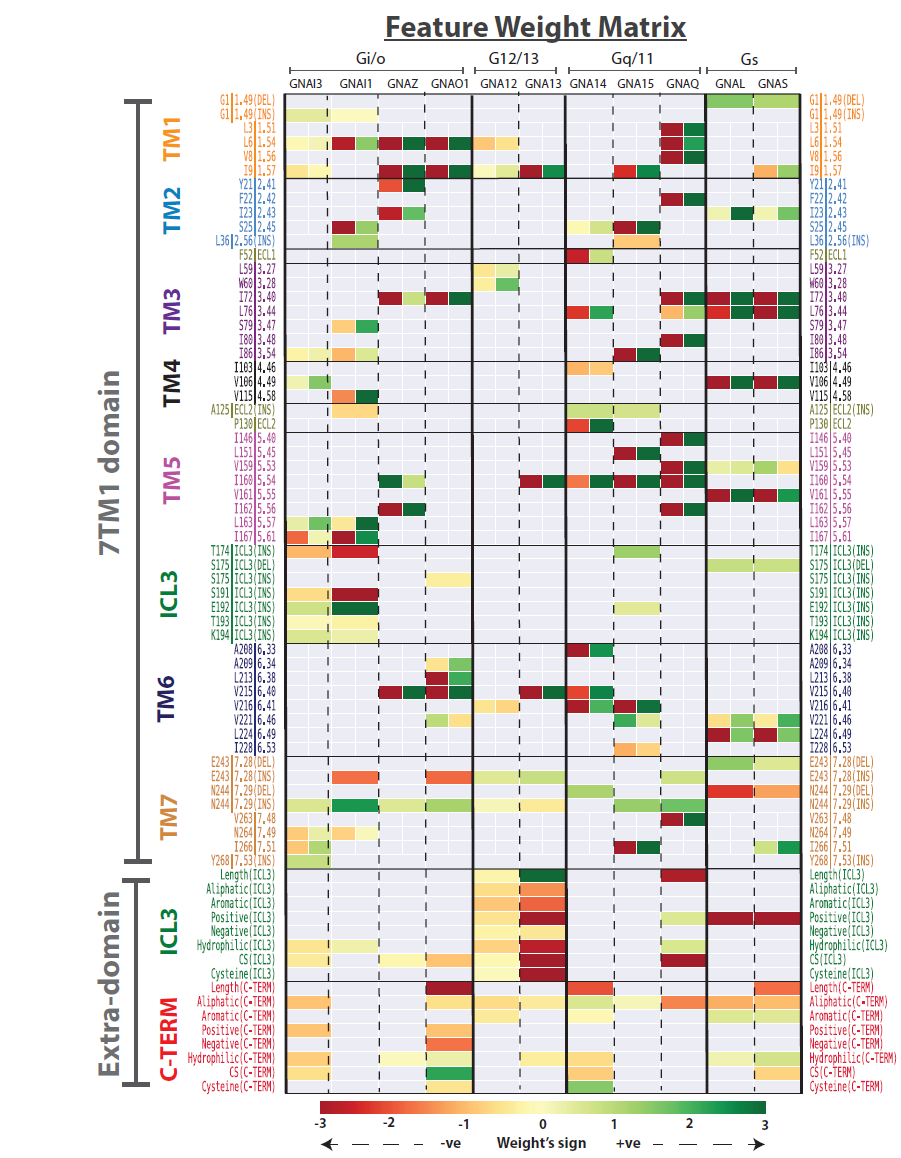 |
Predicting GPCR/G-protein coupling: The GPCR features that are identified as being informative about predicting each of 11 human G-proteins (and subfamilies) |
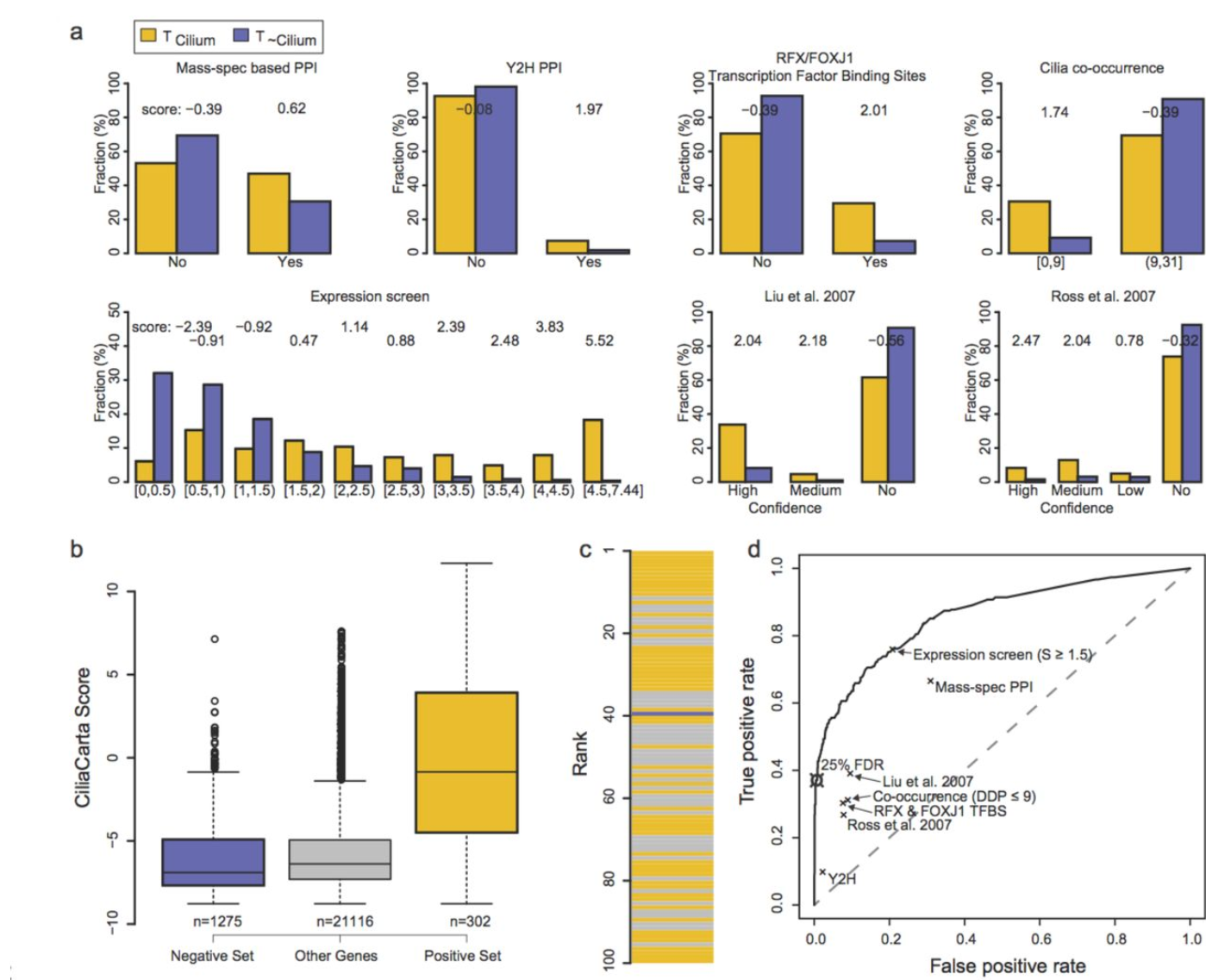 |
Ciliacarta: identifying ciliary genes: Summary of how various independent pieces of evidence predict whether a gene is ciliary or not. |
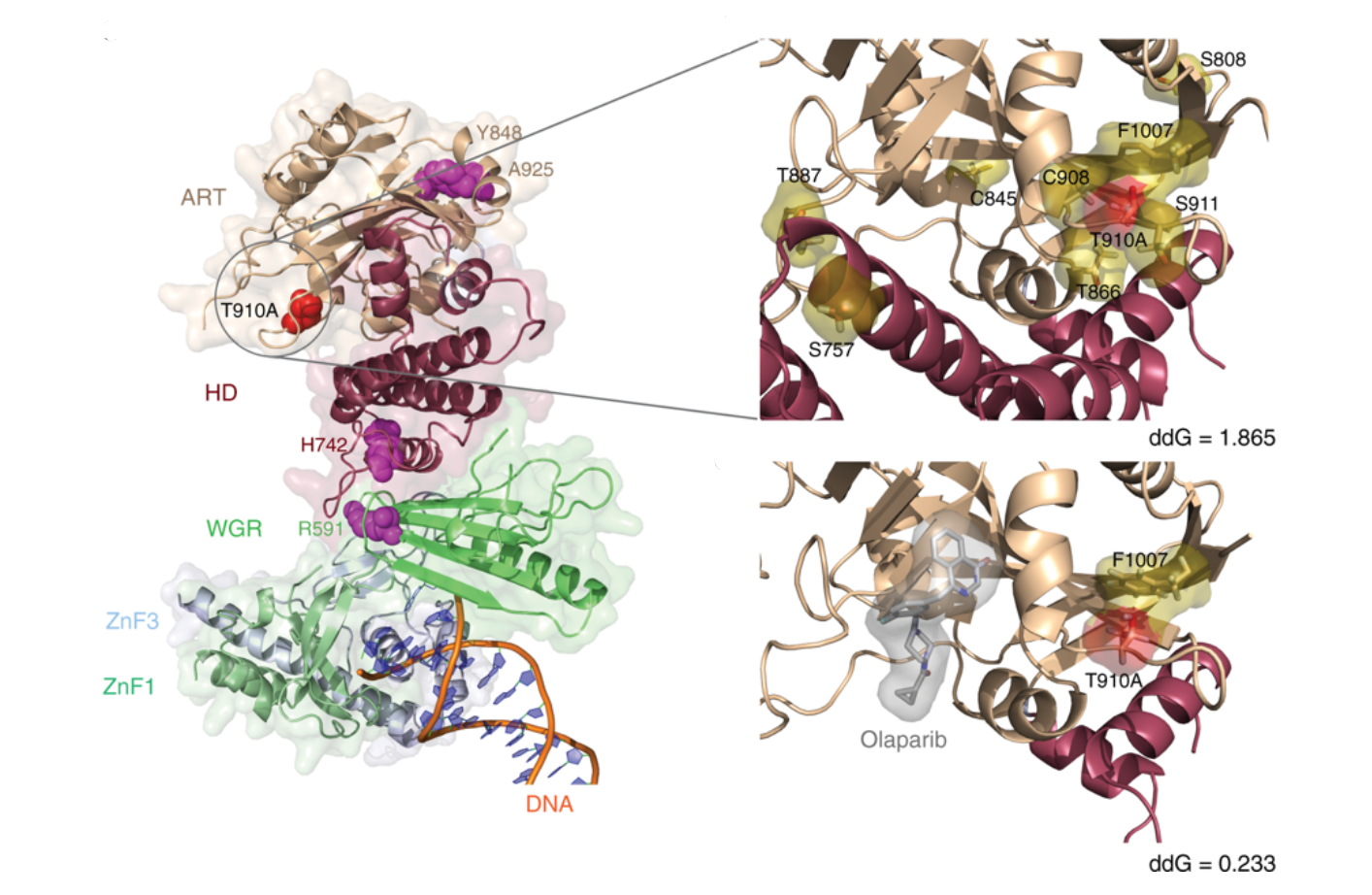 |
New PARP1 mutations in advanced chordomoa: Mutations in PARP1 found in a patient with resistance to an inhibitor and how we estimate they will impact on interaction stability |
 |
An H3K27M mimicking PRC2 inhibitor: Hey, we made the cover. |
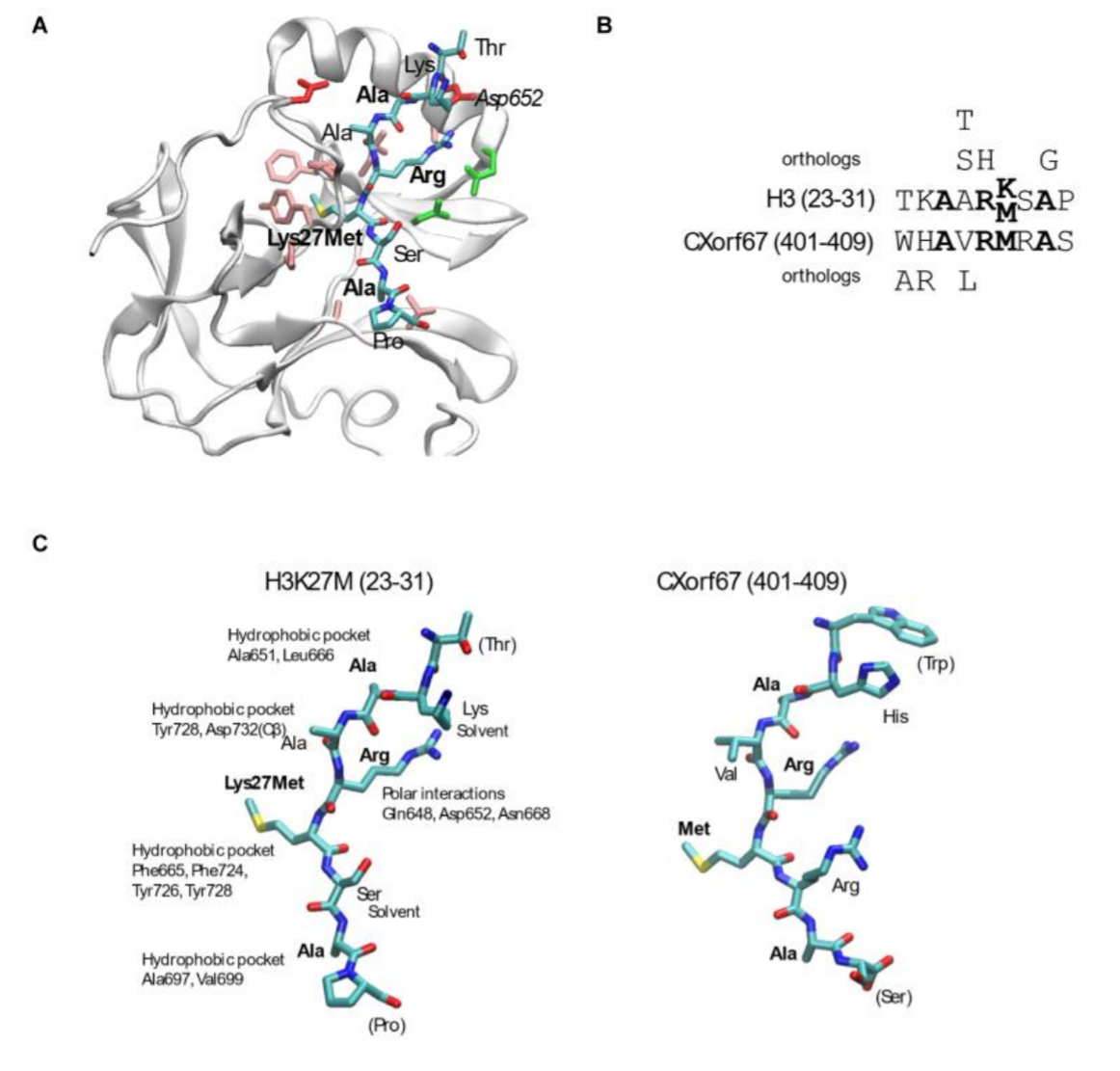 |
An H3K27M mimicking PRC2 inhibitor: The figure shows how we modelled the peptide from CXorf67/EZHIP on the H3K27M EZH2 complex structure and how the two sequences are simimlar in terms of properties |
 |
An H3K27M mimicking PRC2 inhibitor: The conserved C-terminus of CXorf67/EZHIP that resembles the properties of the H3K27M sequence. |
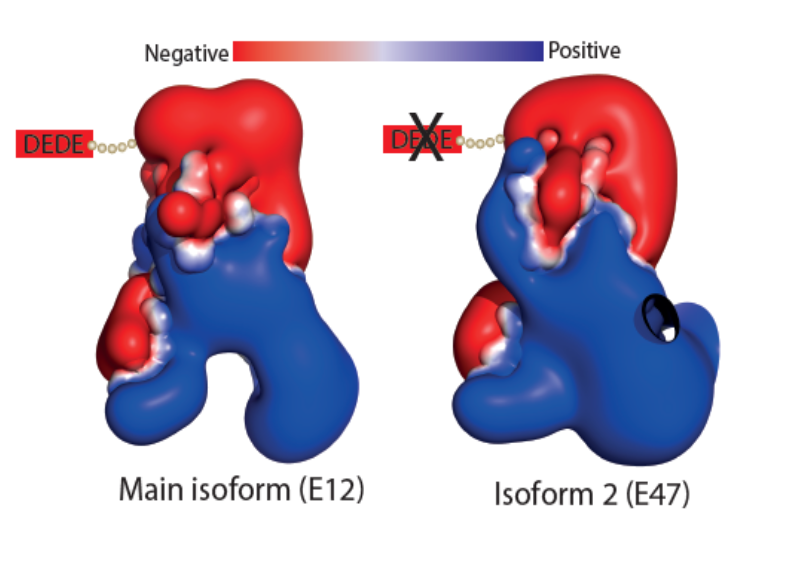 |
The Landscape of mutations in Burkitt Lymphoma: How two isoforms of TCF3 differ in terms of their calculated electrostatic properties. |
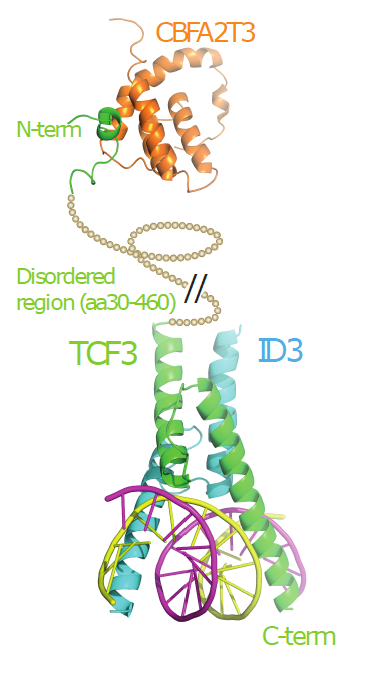 |
The Landscape of mutations in Burkitt Lymphoma: Model of how TCF3 interacts with both ID3 and CBFA2T3 in the context of BL. |
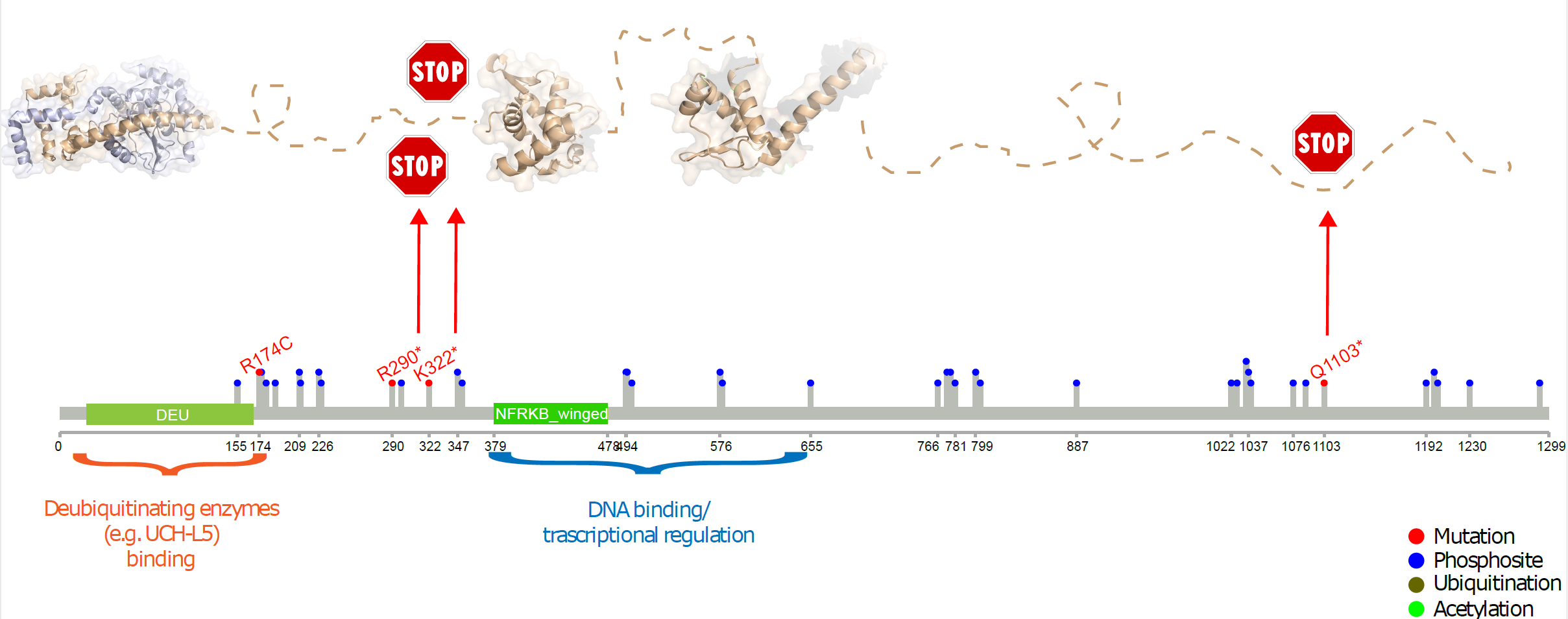 |
NFRKB mutations in Burkitt Lymphoma 11q aberration patients: Summary of observed mutations in NFRKB within this BL patient subset. |
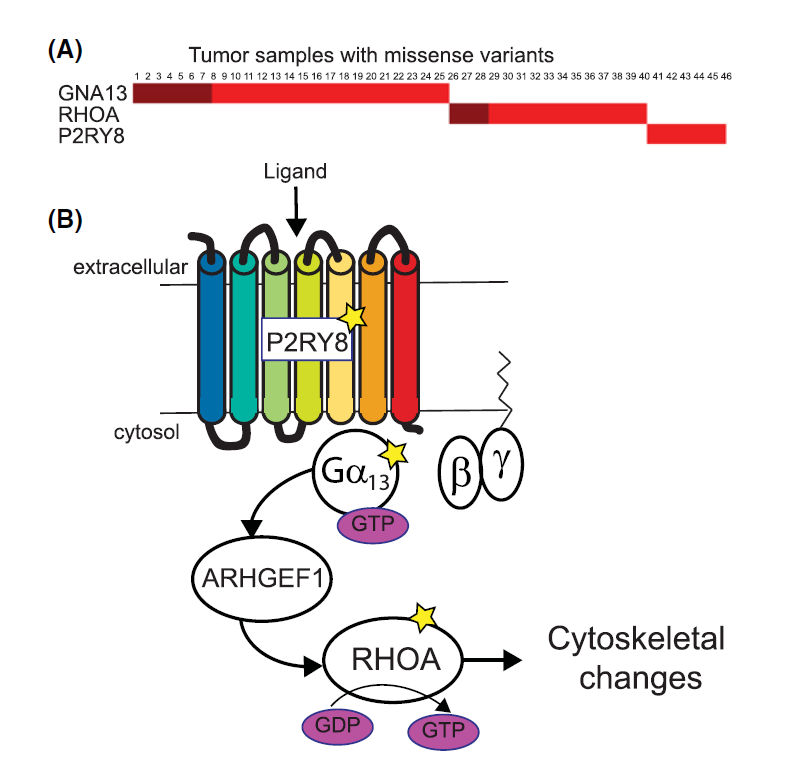 |
Variants affect biological mechanism: Mutually exclusive genes involved in GPCR mediated signalling in lymphoma. |
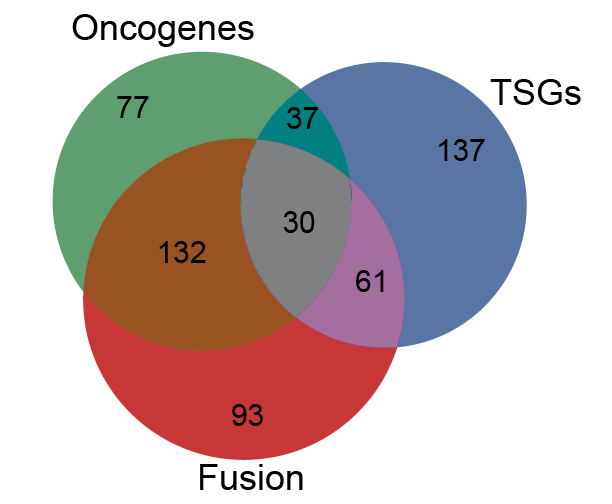 |
Cancer genetics meets mechanism: Overlapping definitions of Oncogenes, Tumor supressors (TSGs) or Fusions in the COSMIC gene census. |
 |
Cancer genetics meets mechanism: Examples of somatic mutations mapped to KRAS and PIK3CA. |
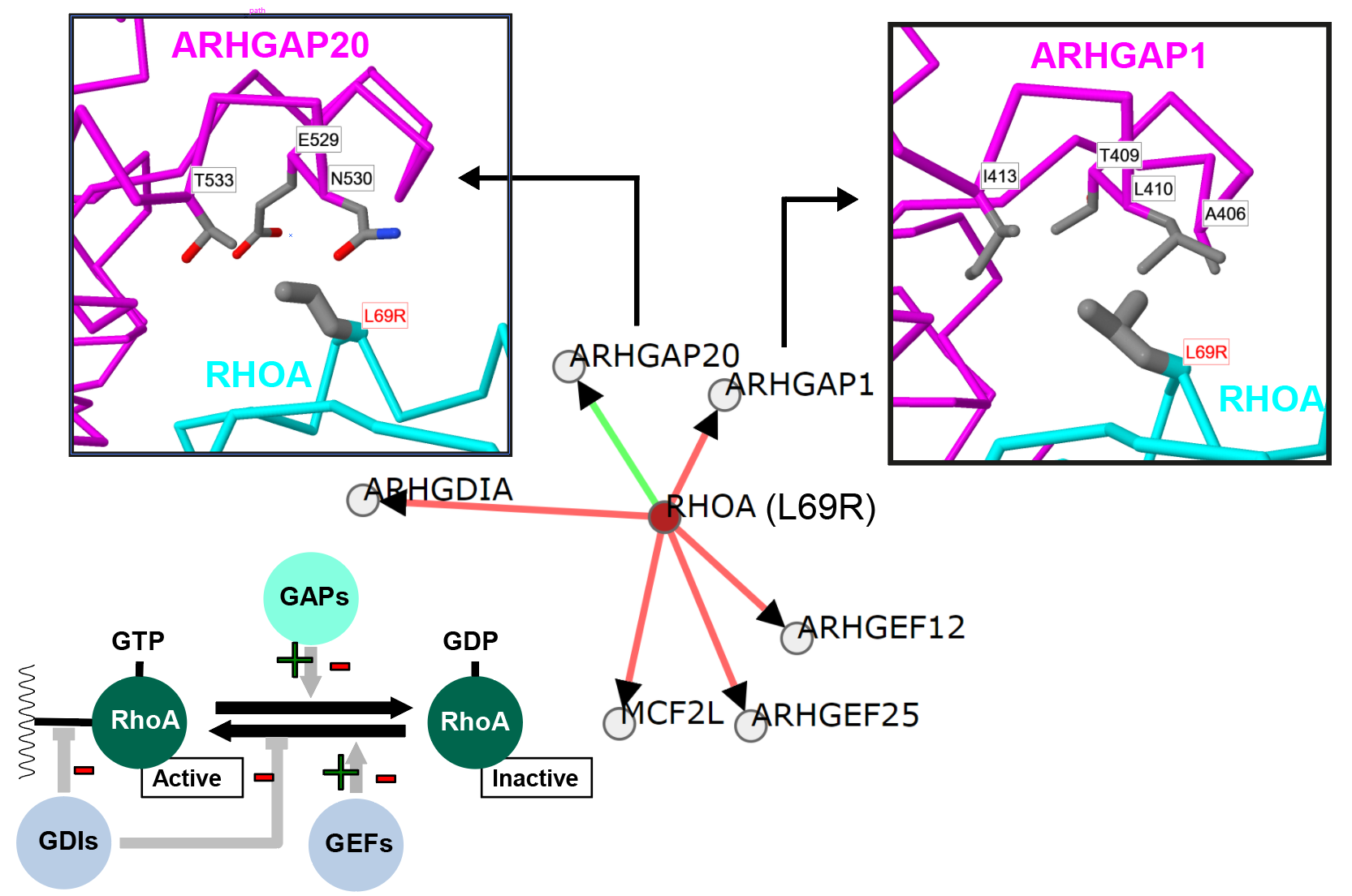 |
Cancer genetics meets mechanism: A Burkitt Lymphoma mutation in RHOA that appears to alter regulation by different GTPase activating (GAP) proteins. |
 |
Genetic variants in cryptic choleostasis: Domain diagram showing location of genetic variants in patients suffereing from cryptic choleostsis. |
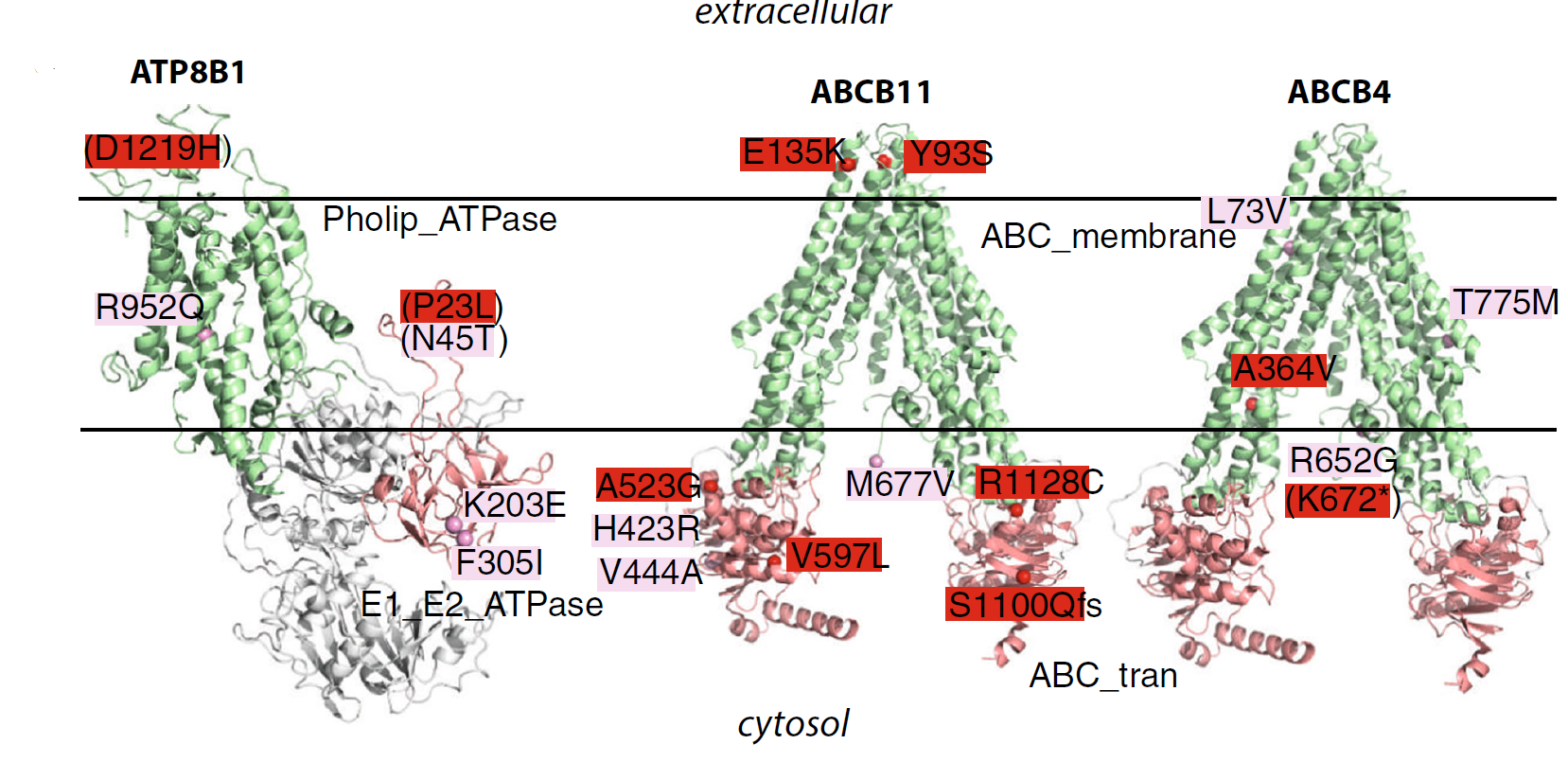 |
Genetic variants in cryptic choleostasis: Models of the four genes studied in patients affected by cryptic choleostsis, showing the location of the mutations. |
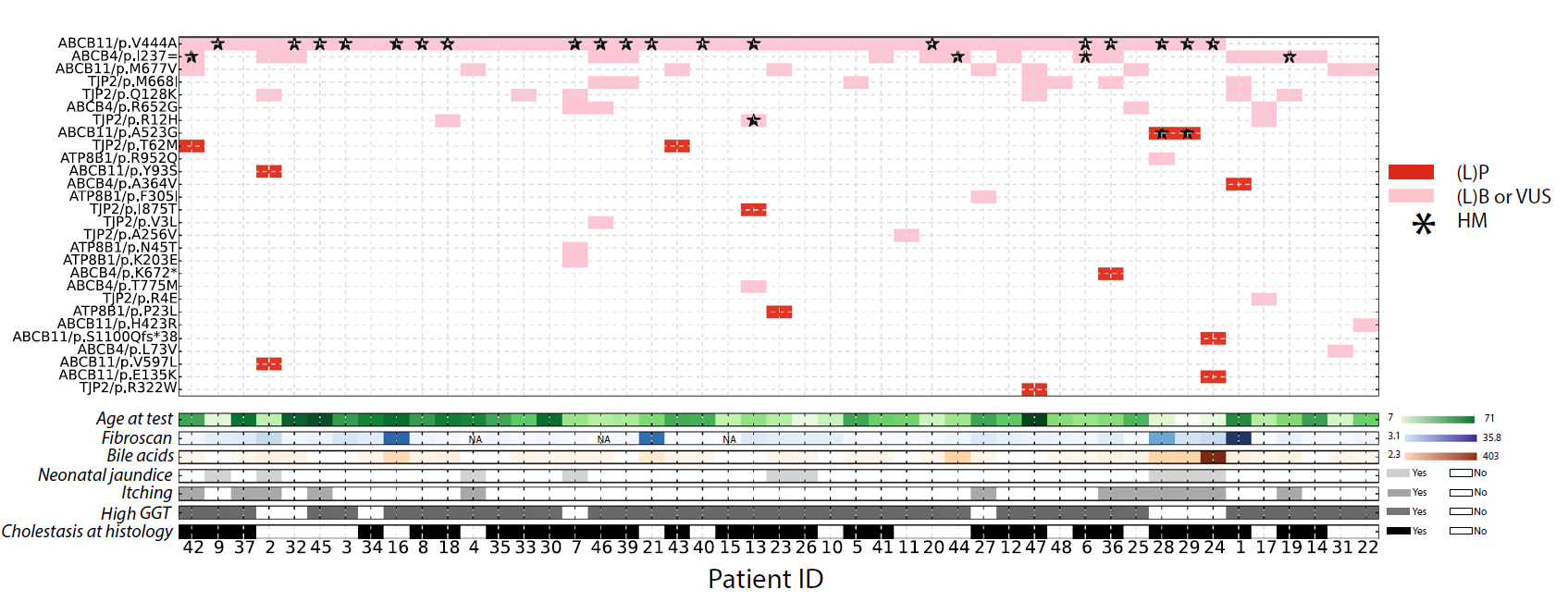 |
Genetic variants in cryptic choleostasis: How the mutations are distributed across genes and patients, and the associated clinical observations. |
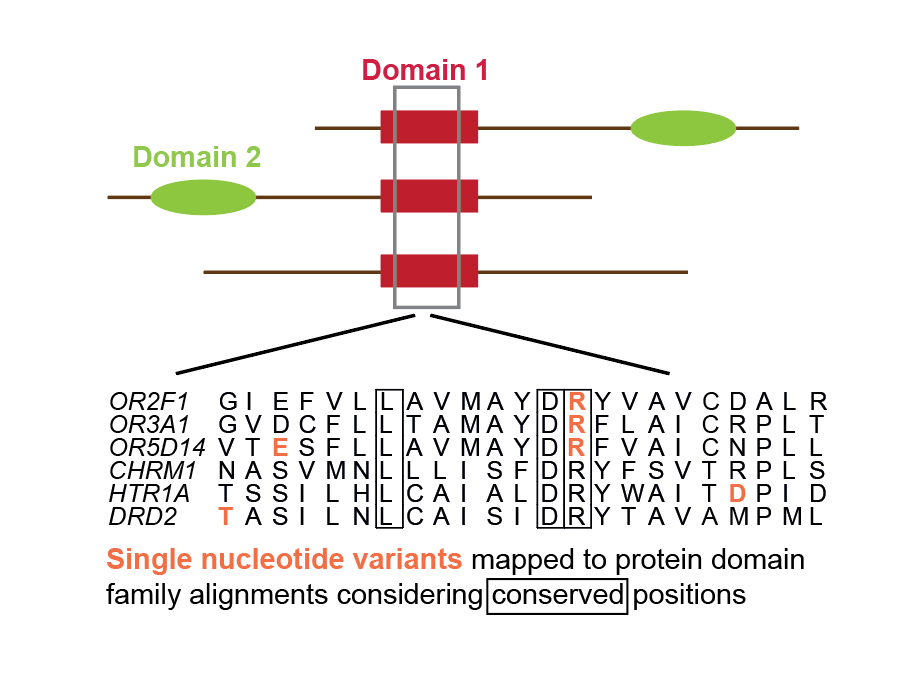 |
Functional genetic variants in healthy genomes: How we identified alignment positions enriched in genetic variants. |
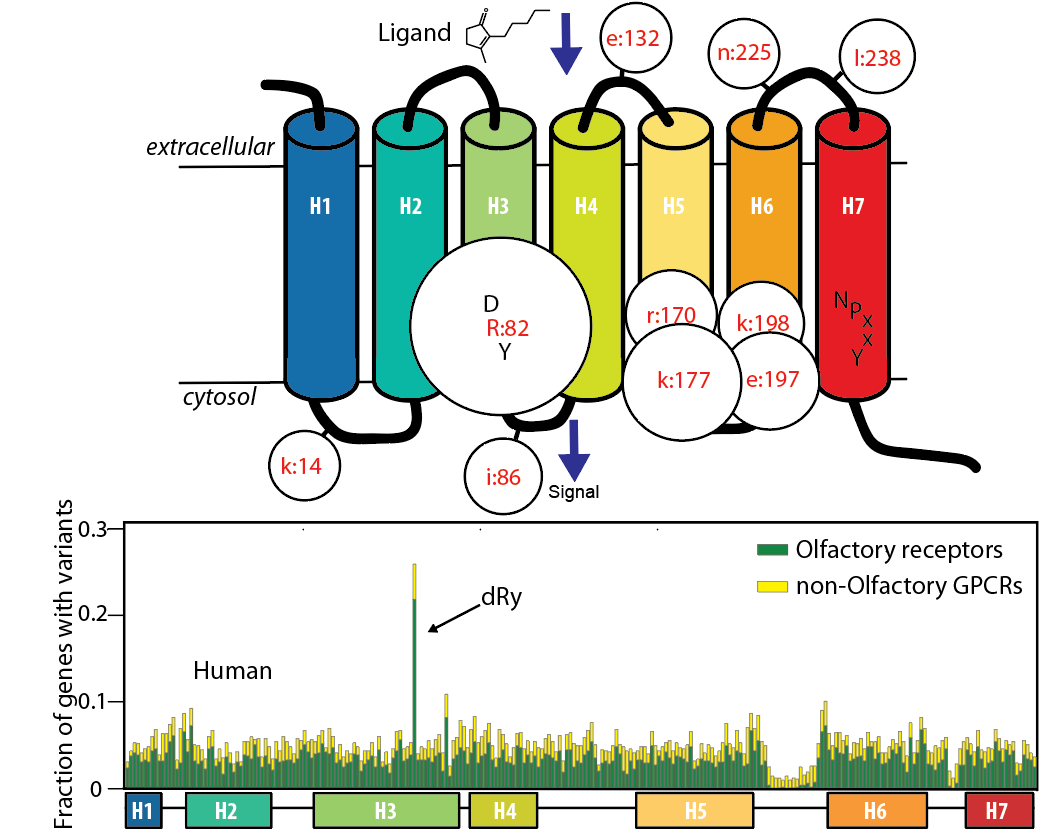 |
Functional genetic variants in healthy genomes: Positions in the GPCR/Olfactory receptor family with significant variant enrichment. |
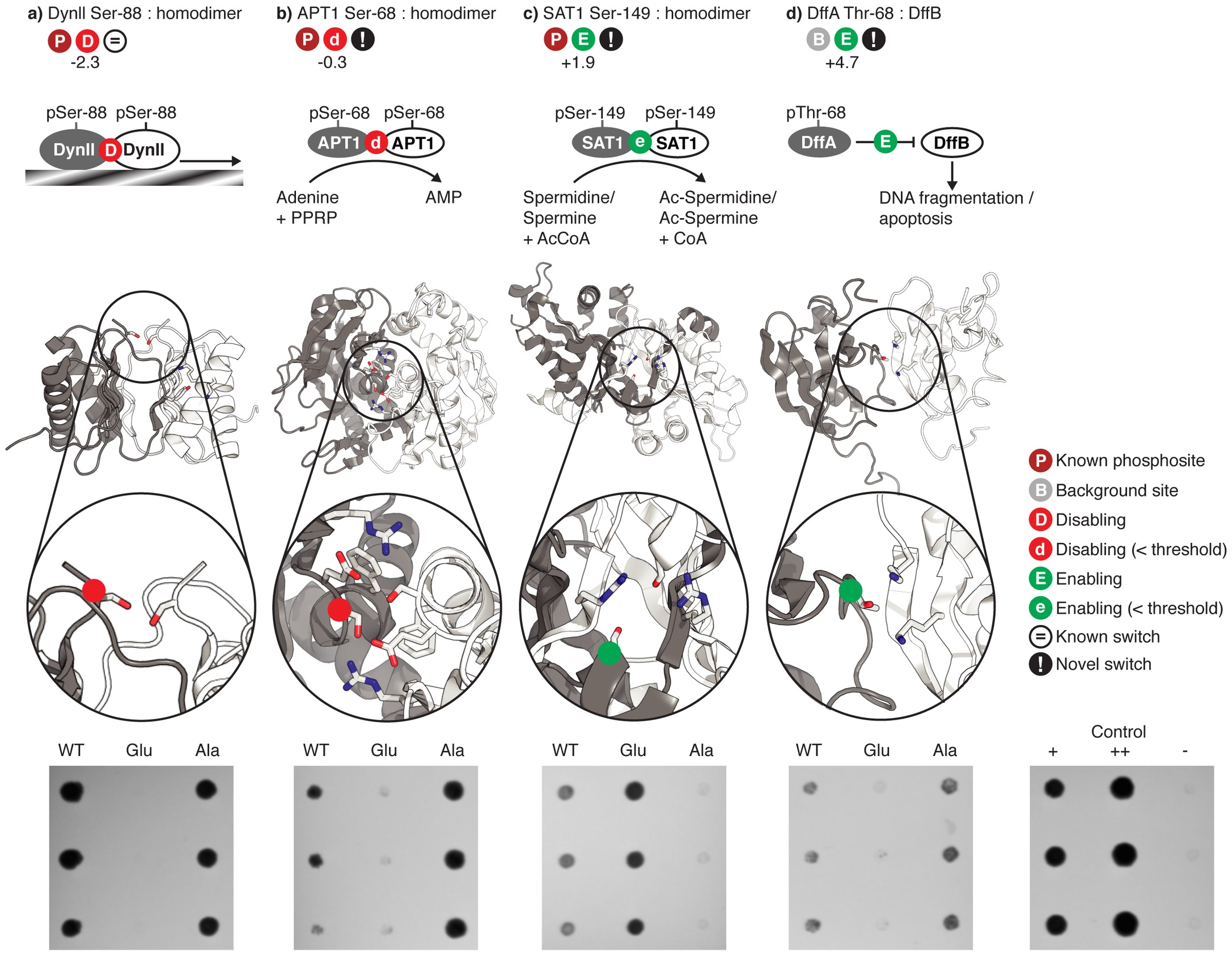 |
Phosphosites targeting interactions: A sample of phosphosites that we predicted to target particular protein-protein interfaces, and which were stesed using the yeast two-hybrid system. |
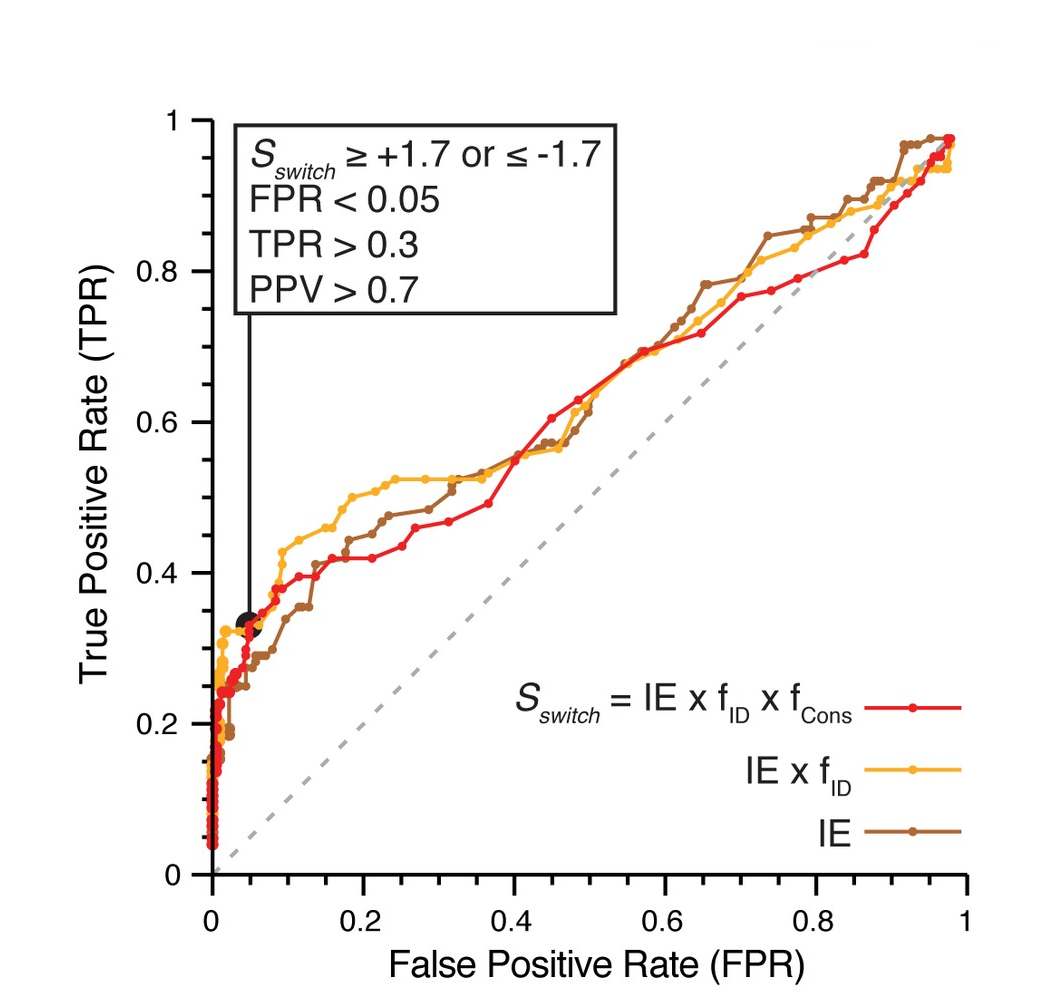 |
Phosphosites targeting interactions: ROC curve showing performance of our interface phosphosite predictor on a benchmark of known sites |
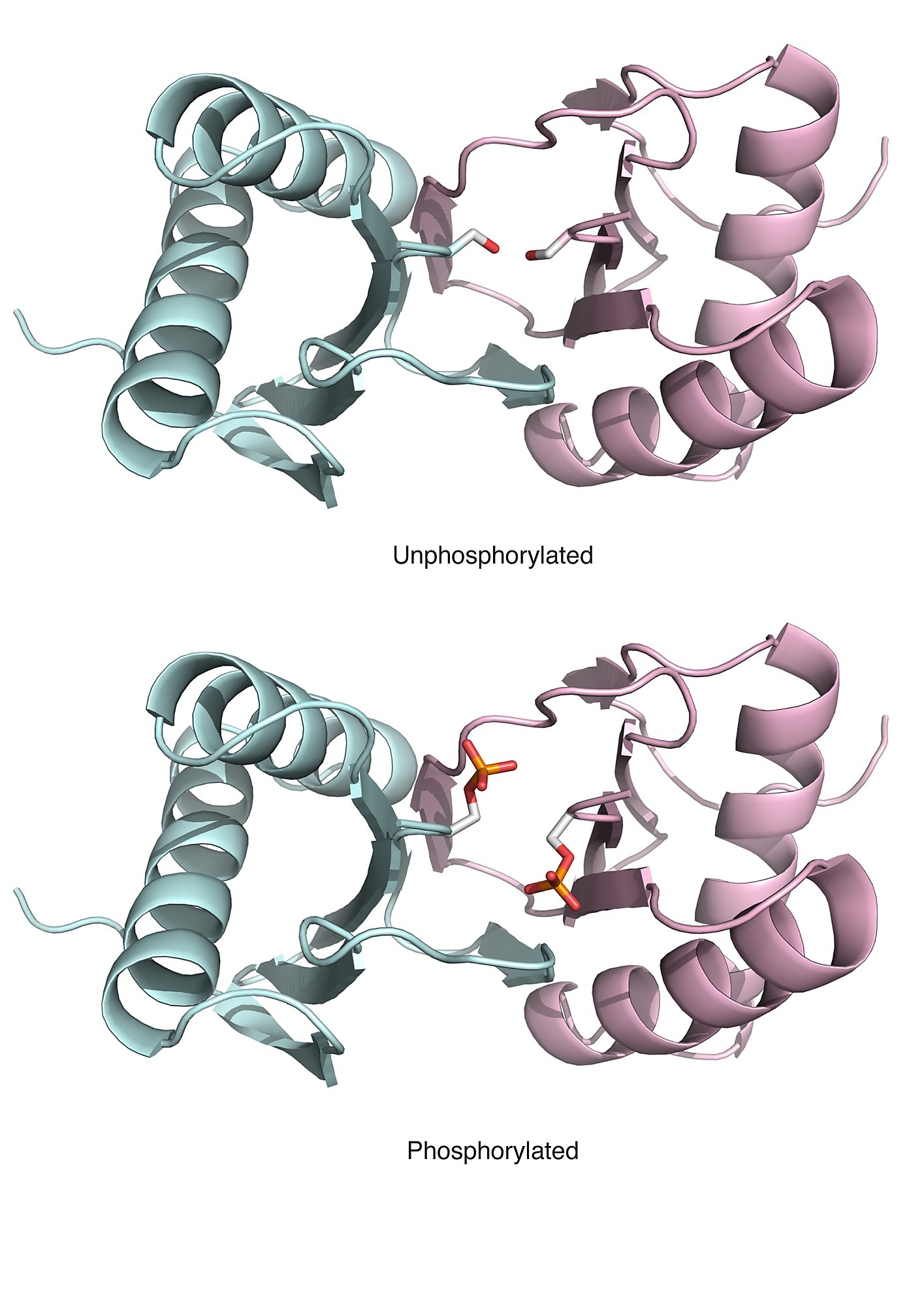 |
Phosphosites targeting interactions: An known example of a phosphosite targeting an interface (Ser-88 in Human Dyenin light chain) |
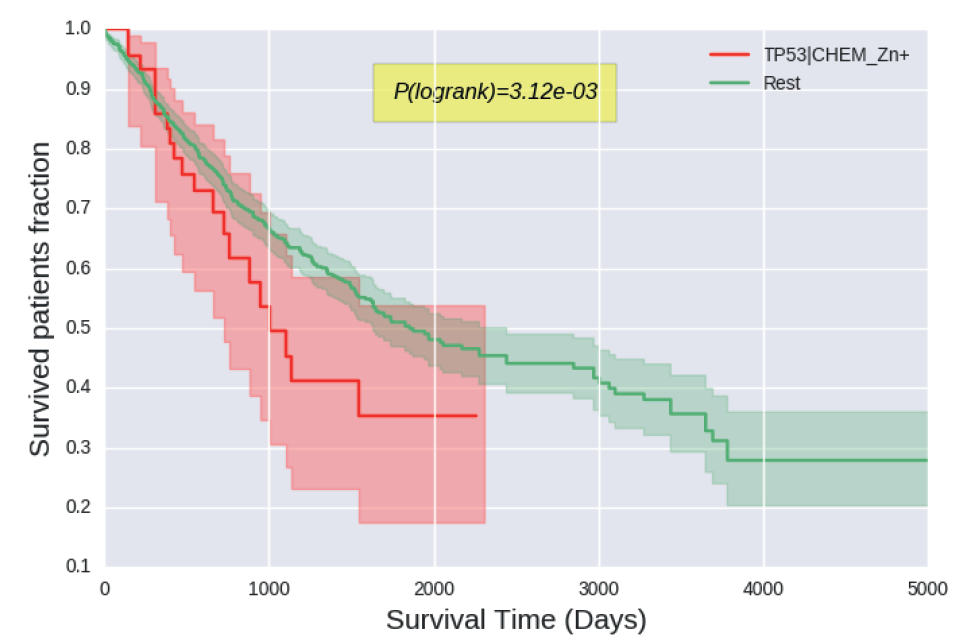 |
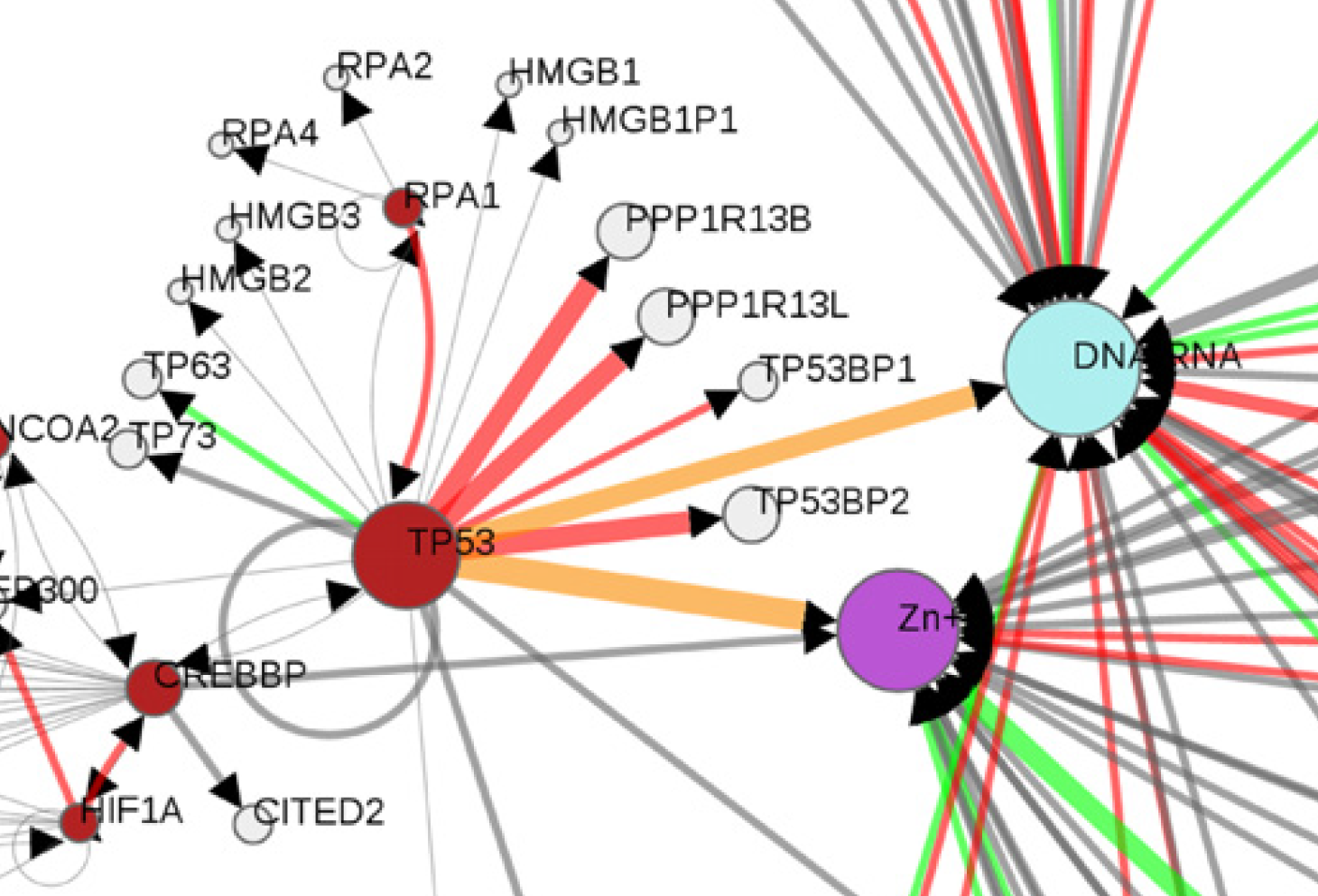
Cancer severity from interaction mechanisms: Figure shows shorter survival across all cancers for tumors with mutations altering the P53-zinc interface (red) compared to others (green). |
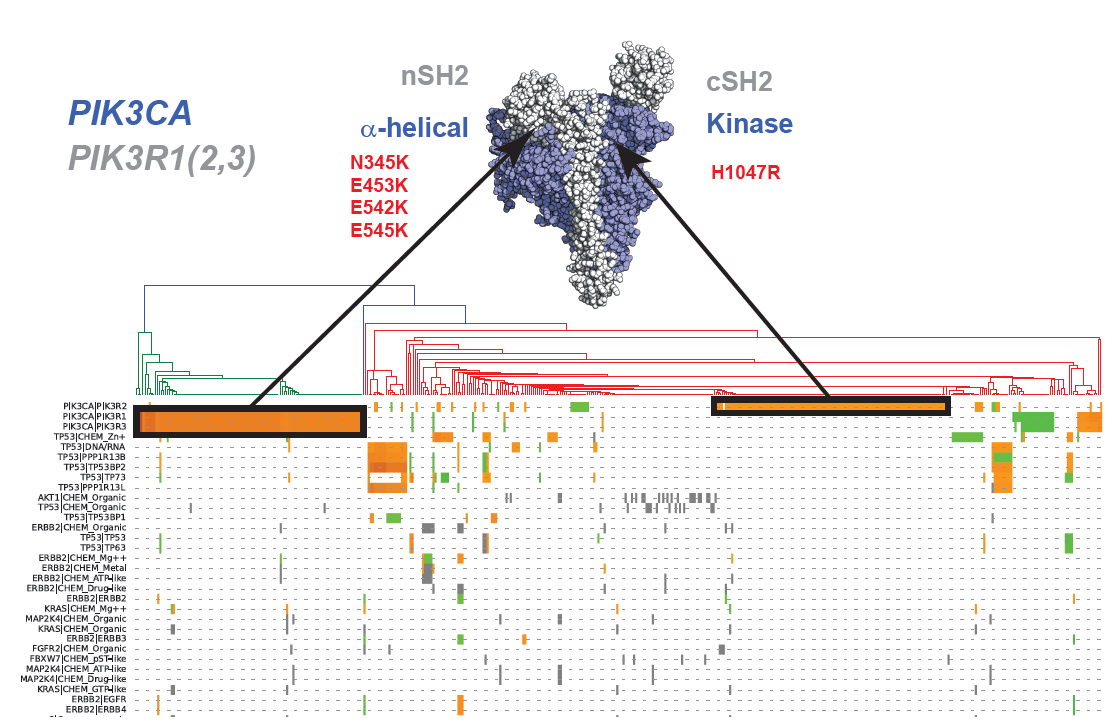 |
Cancer severity from interaction mechanisms: Figure shows distinct populations within Breast carcinoma according to different mutations at the PIK3CA/PIK3R1 interface. |
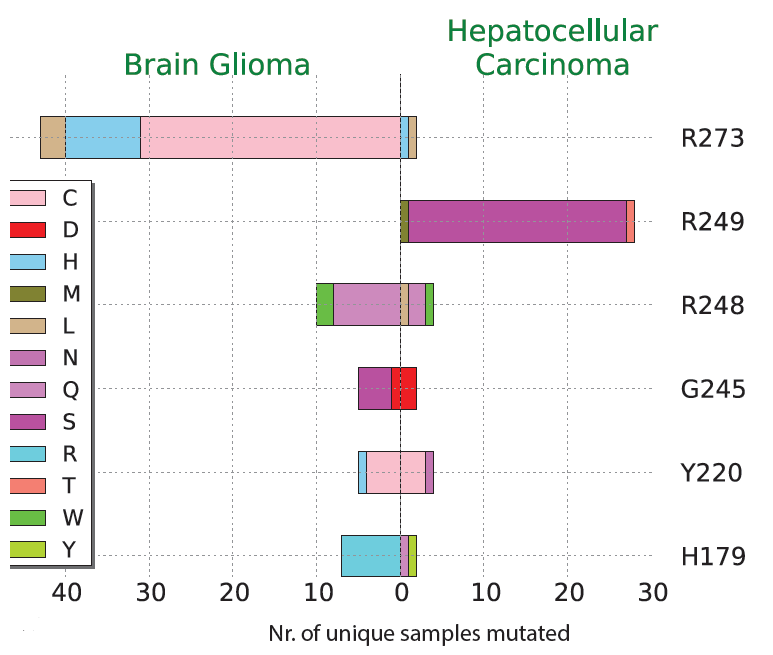 |
Cancer severity from interaction mechanisms: Figure shows differences in TP53 mutation preference in the two cancer shown. For example, Arginine (R) 273 is a preferred site in Brain Glioma, and the majority of changes are to Cysteine (C). |
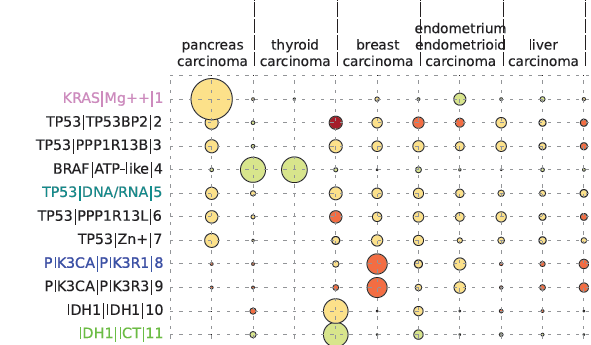 |
Cancer severity from interaction mechanisms: Figure shows how particular interfaces are targetted in different cancers. The size of the circle indicates how many mutations are seen, and the colour indicates whether the mutations are mostly disabling (red) or enabling (green). |
 |
Modelling of FFV gag protein: Figure shows mutations affecting the Gag–Elp interface, central beta-sheet affect budding or Gag folding. |
 |
Ciliary landscape: Figure shows the effect knocking down components of proteins where mutations lead to 3M syndrome have on cilia in mpkCCD cells. We hypothesized that 3M Syndrome is a previously undescribed ciliary disease. |
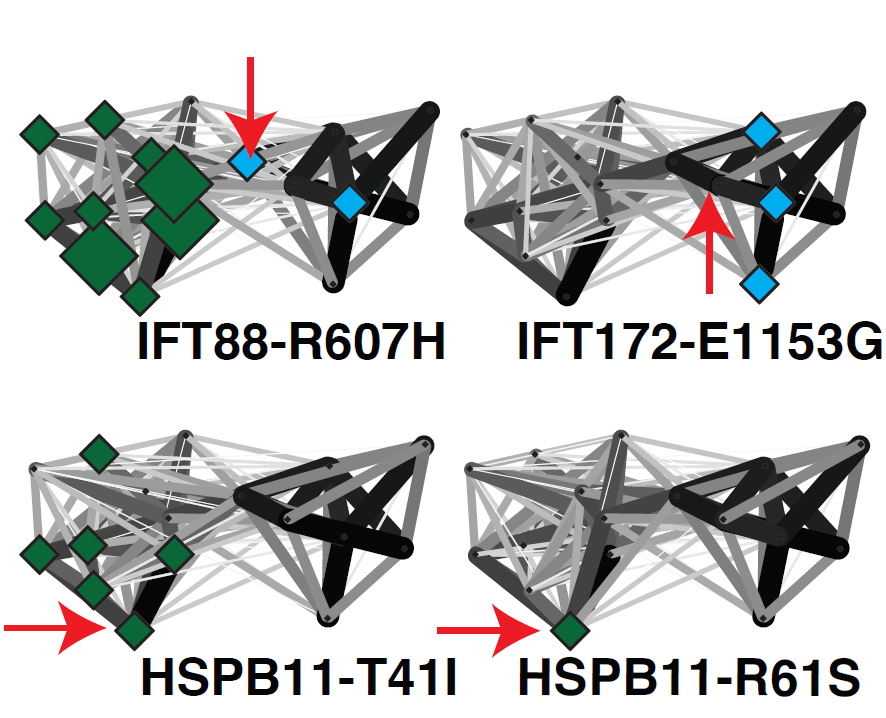 |
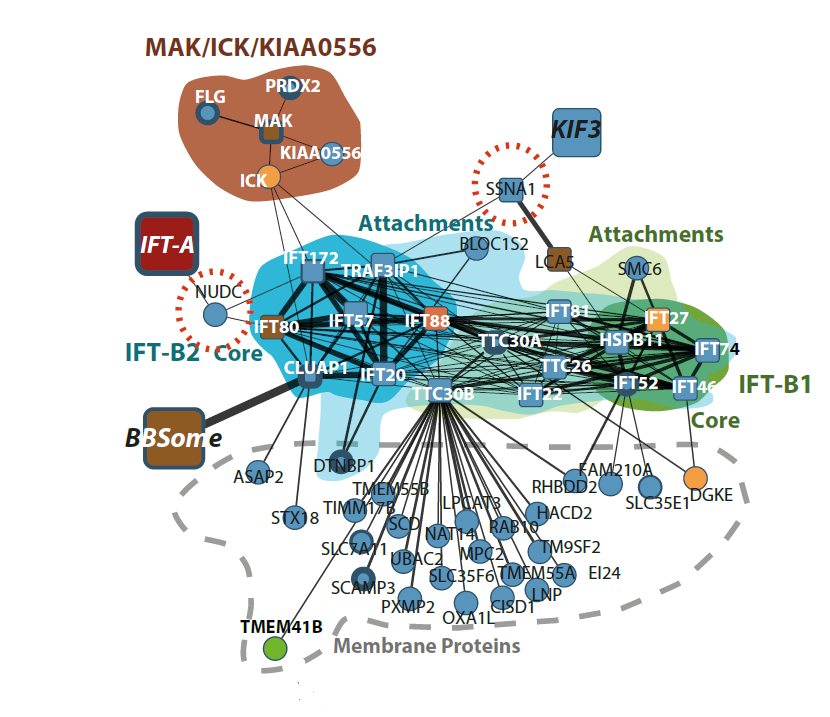
Ciliary landscape: Figure shows the effect of variants in patients suffereing sever ciliary diseases and how they affect specific sub-complexes and interaction within the intraflagellar transport complex B. |
 |
Ciliary landscape: Figure shows the architecture of the intraflagellar transport complex B deduced by our Socioaffinity metric. |
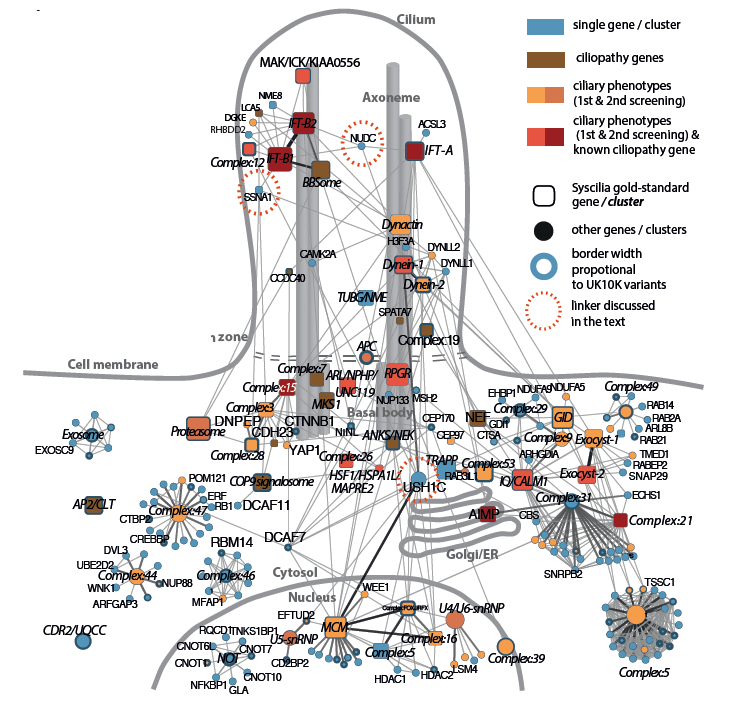 |
Ciliary landscape: Figure shows an overview of the complexes and interaction within the human primary cilium uncovered by Interaction Proteomics. |
 |
Mechismo: Figure shows a graphical representation of mutations and their effects on biomolecular interactions within Pancreatic cancer, including whether mutations enhance (green) or diminish (red) particular interactions. Generated using the Mechismo server. |
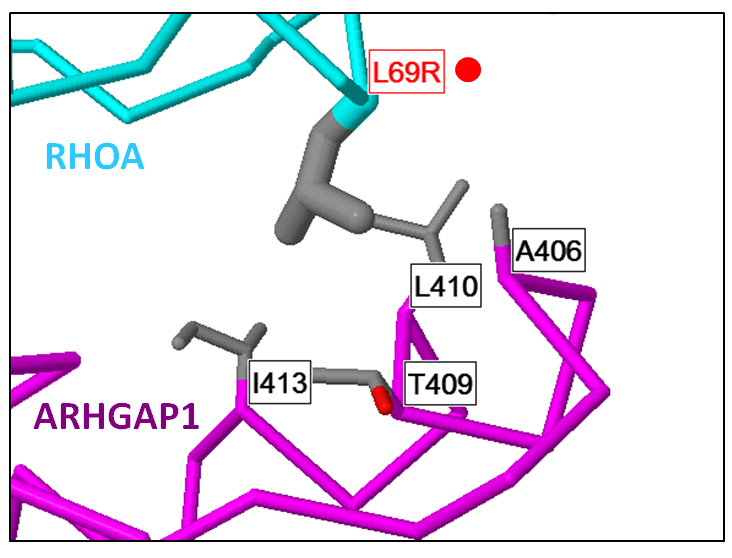 |
Mechismo: Figure shows an example of a mutation within RhoA (L69R) thought to enhance an interaction with ARHGAP1 within Burkitt's lymphoma. Generated using the Mechismo server. |
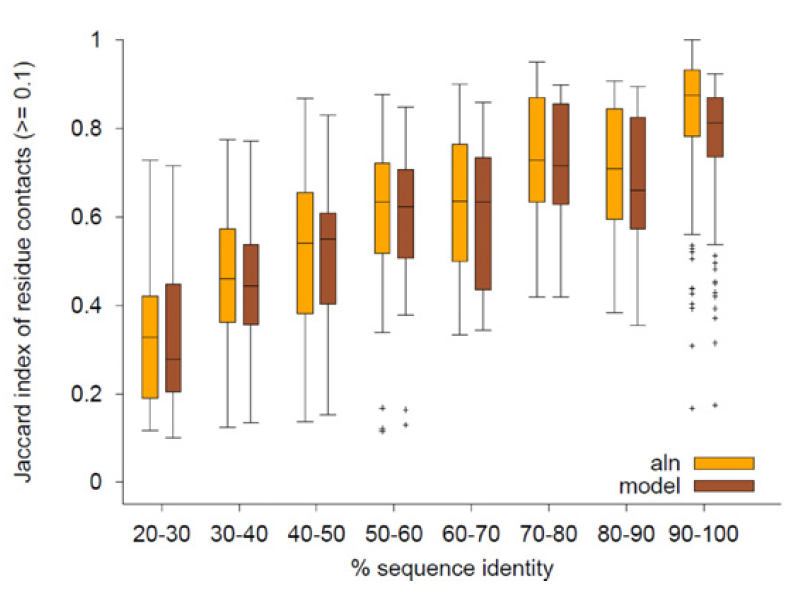 |
Mechismo: Figure shows a comparison of automated modelling techniques (model) to a simple strategy of looking at an alignment to template structure (aln) in terms of the fraction of residue contacts reproduced (as a function of sequence identity). |
 |
siRNA to protein-interaction mechanism: Figure shows how interactions within the IL2 signalling pathway are reproduced by the HIPPIE approach to deduece protein-interaction directionality using siRNA screen data. |
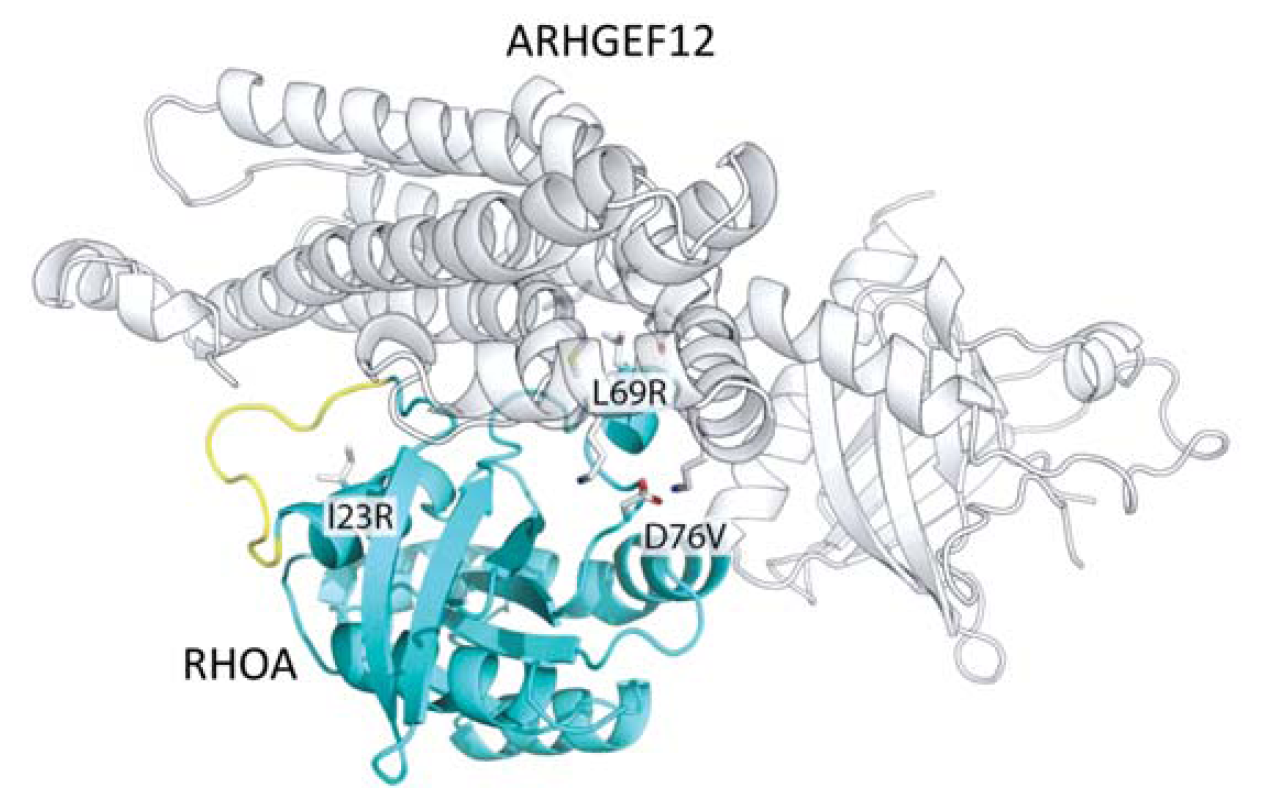 |
RhoA mutations in Burkitt's lymphoma: Figure shows the location of key mutations within Burkitt's lymphoma that lie and the interface with one of its modulators (ARHGEF12) |
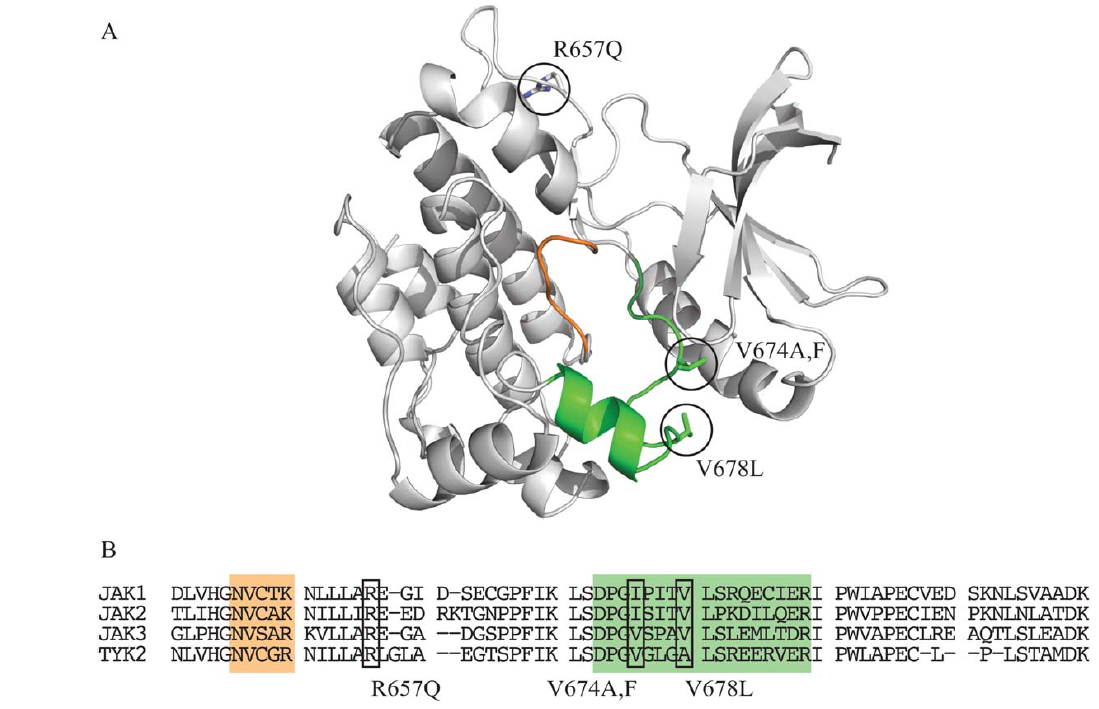 |
JAK3 mutations in T-cell prolymphocytic leukemia: Figure shows the location of key mutations in this cancer thought to affect JAK3 function in particular ways. |
 |
Structure of Argonaute proteins by DNA shuffling: Figure shows the location of functional regions in modesl of Ago2 and Ago3 deduced by DNA shuffling techniques. |
 |
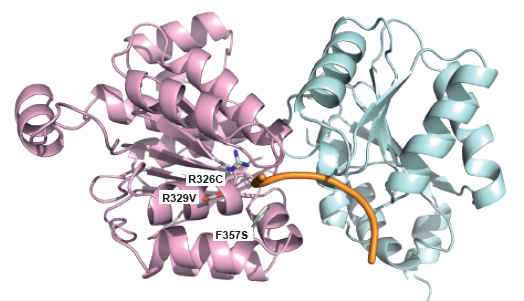
Mutations in ID3/TCF4 associated with Burkitt's Lymphoma: Figure shows the location of mutations observed in patients suffering from from Burkitt's Lymphoma at the dimer-interface and DNA binding site of TCF3 |
 |
Mutations within Medulloblastoma: Figure shows the location of mutations observed in patients suffering from Medulloblastoma within the DNA binding site of DDX3X |
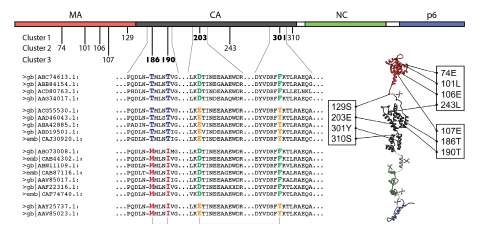 |
Correlated mutations within HIV Gag: Figure shows the locations of predicted correlated positions within HIV Gag |
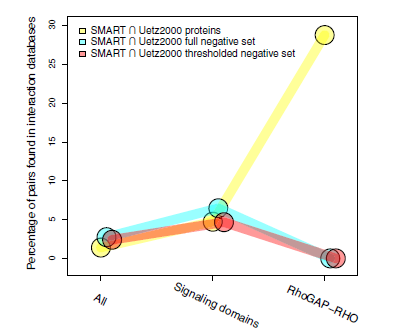 |
Defining negative protein-protein interact datasets: Figure shows how the rational selection of negatives improves the prediction performance for specific biological contexts |
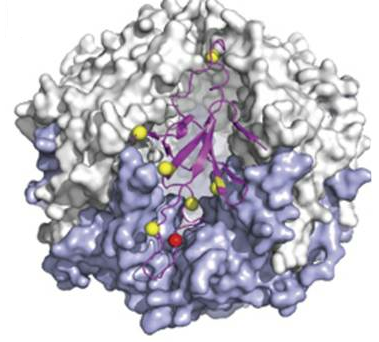 |
Modification in mycoplasma pneumonia: Figure shows the location of phosphorylation (red) and acetylation (yellow) events in mycoplasma that occur at the interface of a GroES oligomer |
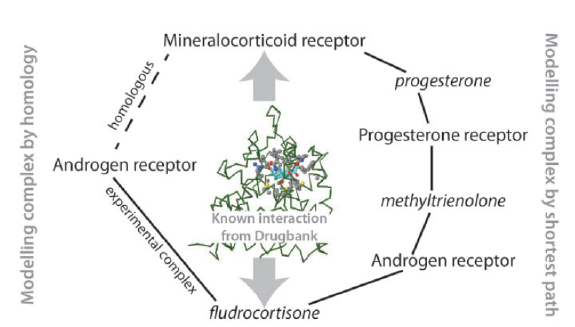 |
Predicting drug-protein interactions: Figure shows how our method for predicting protein-chemical interactions can link proteins and chemicals via a series of protein/chemical superimpositions. |
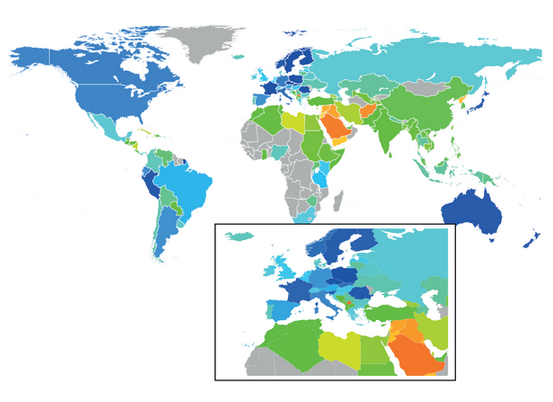 |
Wikipedia diputes reflect geopolitical instability: Mercator projection with countries coloured according to a dispute-index heat map (blue: few disputes; red: many disputes). |
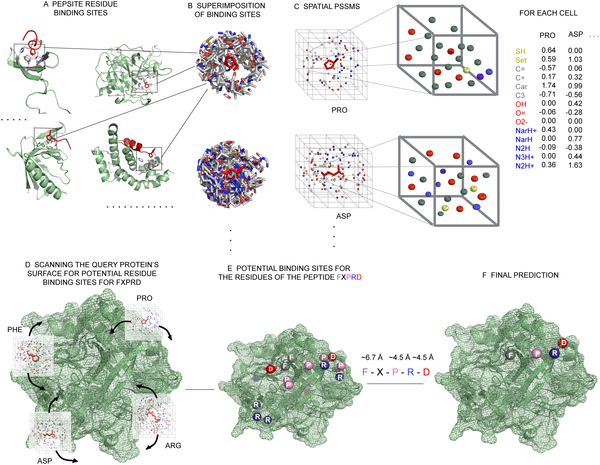 |
PepSite: A schematic detailing our PepSite approach for finding peptide binding sites on protein surfaces. |
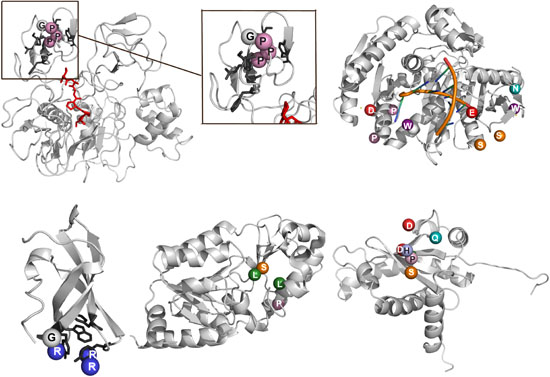 |
PepSite: A demonstration of some of the peptide binding sites predicted correctly using our program PepSite. |
 |
Mycoplasma modelling: A figure showing modelling details for the Mycoplasma pneumonia RNA polymerase |
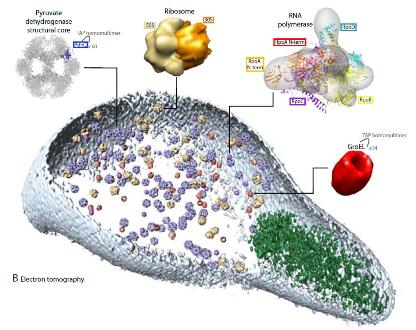 |
Mycoplasma modelling: Figure showing how we fit our models from Mycoplasma pneumoniae into a Cryoelectron tomogram for the bacteria. |
 |
WD40-peptide interactions: Figure from our review on the subject showing the location of peptide binding sites on this very promiscuous protein family |





















































