 |
Mechnetor A tool to predict interaction mechanism and interrogate mutations, variants, PTMs in proteins
From Gonzalez-Sanchez et al Nucl Acids Res, 2021. |
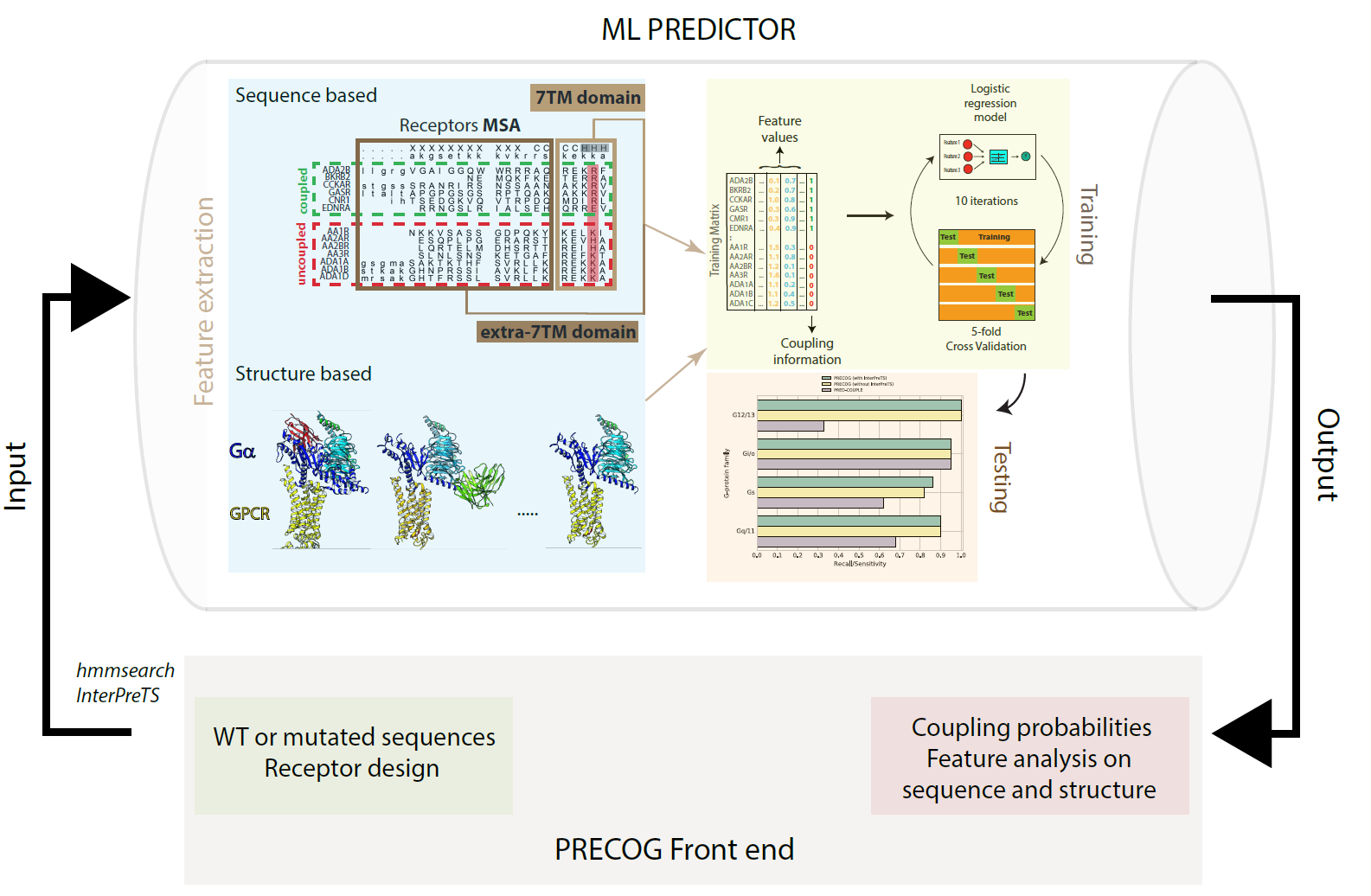 |
PRECOG A tool to predict how GPCRs couple (signal) through G-proteins.
From Singh et al Nucl Acids Res, 2019. |
 |
Syscilia landscape This tool lets you interrogate data from our collaborative project on the Ciliary proteome, where we identified nearly 5000 interactions and over 50 complexes, involving more than 1000 proteins in the cilia.
From Boldt, van Reeuwijk, Lu, Koutroumpas et al Nat Commun, 2016. |
 |
GetGo A fast means of looking at enrichment of human genes in GO terms, pathways, and other clusters. We developed this mostly as other servers were too slow, and we wanted to add some datasets and include a better measure of depletion (i.e. as opposed to just enrichment). This is very crude, be warned.
From Boldt, van Reeuwijk, Lu, Koutroumpas et al Nat Commun, 2016. |
 |
Mechismo Mechismo is a tool that lets you find potential mechanisms for how proteins interact with other molecules and more importantly for how any mutations or post-translational modifications that might affect these interactions, and consequently an entire biological system.
From Betts et al, Nucl Acids Res, 2014. |
 |
ProtChemSI A database of protein-chemical interactions predicted by structure superimpositions: if you are interested in chemicals that might bind a particular protein or vice versa, then check it out.
From Kalinina et al, PLoS Comp Biol, 2011. |
 |
PepSite A method to predict the location of peptide binding sites on protein surfaces:
if you have a protein structure and a known or putative peptide that binds to it, pepsite is for you. From Petsalaki et al, PLoS Comp Biol, 2009. |
 |
Linear Motif Discovery (old version here) From Neduva et al PLoS Biology, 2005; Neduva et al Nucl. Acids Res, 2006; Petslaki et al, PLoS Comput Biol, 2009; Trabuco et al, Methods 2012. |
 |
3D-ID: Interacting domains in three-dimensions From Stein et al Nucl. Acids Res, 33, D413-4178, 2005 |
 |
3SOM: 3D Surface Overlap Maximization From Ceulemans & Russell J. Mol. Biol., 338, 783-793, 2004 |
 |
Drosophila MicroRNA target predictions From Stark et al PLoS Biology, 1, 397-409, 2003 |
 |
DisEMBL: Intrinsic Protein Disorder Prediction From Linding et al, Structure, 11, 1453-1459, 2004 |
 |
PINTS: Patterns In Non-homologous Tertiary Structures Based on Stark et al, J. Mol. Biol., 326, 1307-1316, 2003. |
 |
Interaction Prediction through Tertiary Structure Based on Aloy & Russell, Proc Natl Acad Sci USA, 99 : 5896-5901, 2002. |
 |
Prediction Of Unknown Sub-types (PROUST) II Based on Hannenhalli & Russell, JMB, 303, 61-76, 2000. |
 |
Properties of the amino acids See also Betts & Russell, Amino acid properties and consequences of subsitutions, Bioinformatics for Geneticists, M.Barnes & Ian Gray eds, Wiley, 2002. |



